Publications
2025

A new molecular seed assay to predictUstilago nudafield infection levels
Cold Spring Harbor Laboratory
·
27 Jun 2025
·
doi:10.1101/2025.06.25.661478
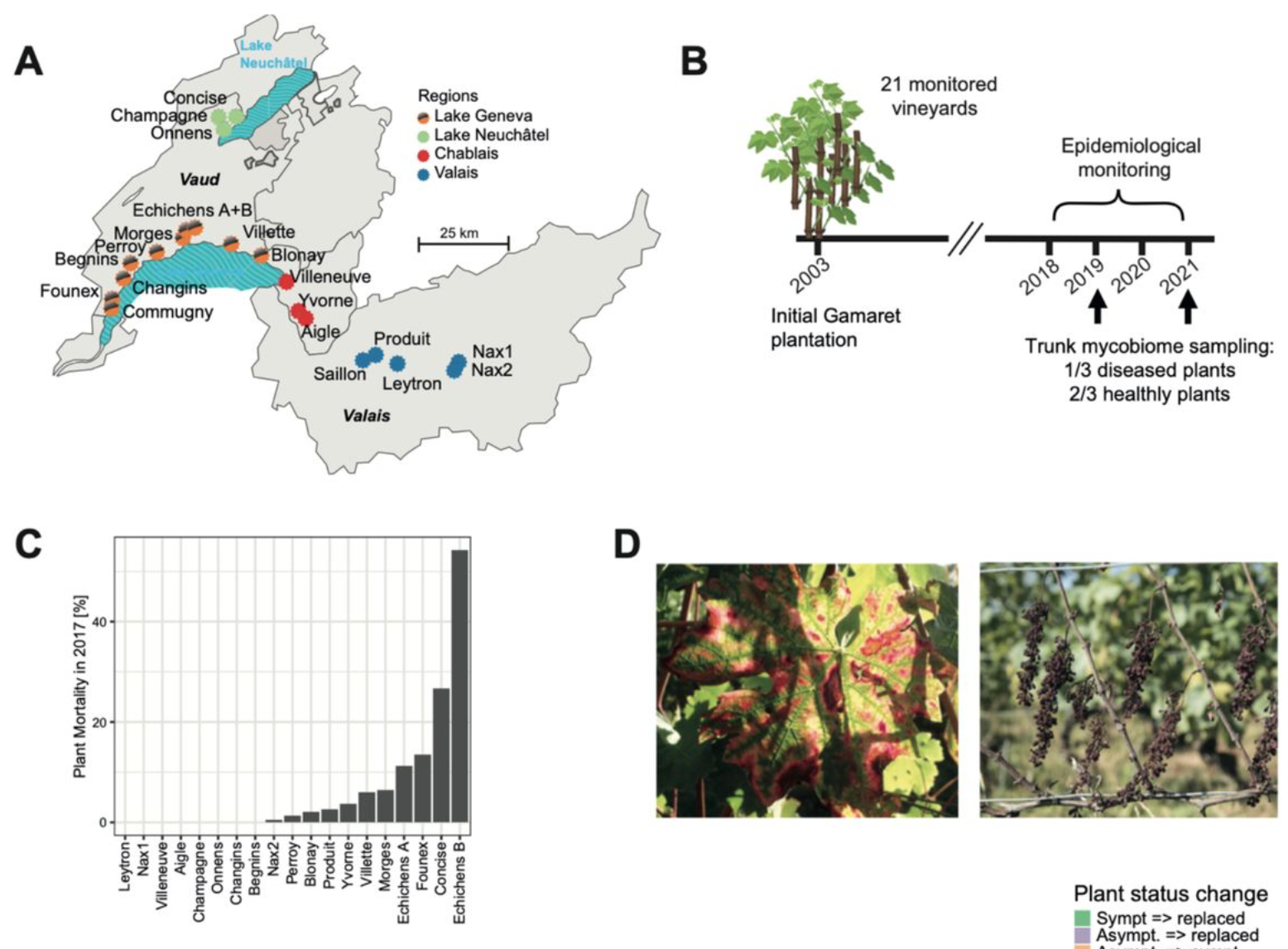
Landscape-scale endophytic community analyses in replicated grapevine stands reveal that dieback disease is unlikely to be caused by specific fungal communities
Applied and Environmental Microbiology
·
20 Jun 2025
·
doi:10.1128/aem.00782-25
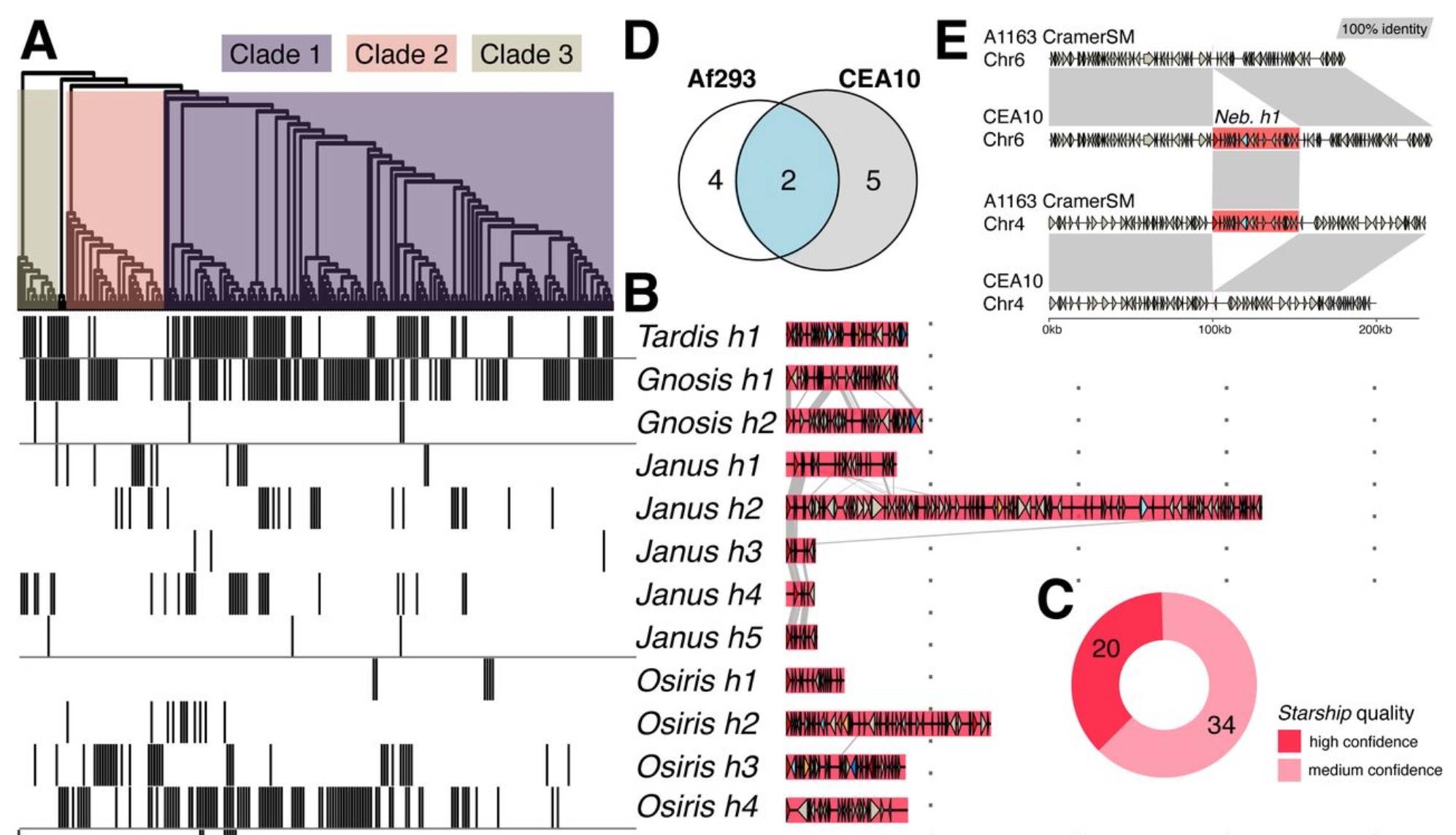
Giant transposons promote strain heterogeneity in a major fungal pathogen
mBio
·
11 Jun 2025
·
doi:10.1128/mbio.01092-25
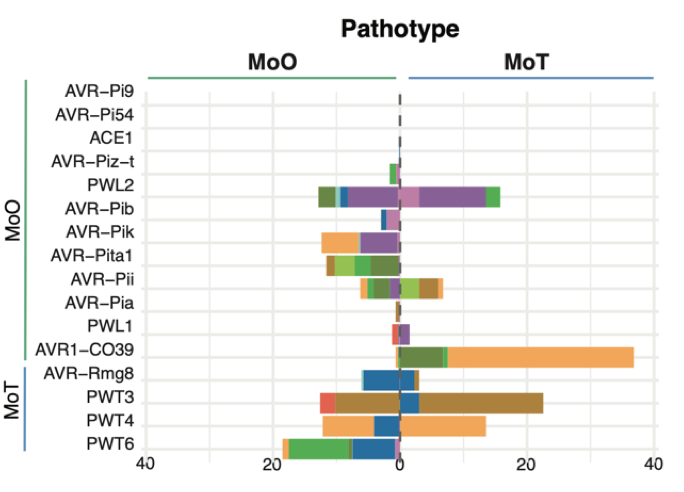
Transposable elements create distinct genomic niches for effector evolution amongMagnaporthe oryzaelineages
Cold Spring Harbor Laboratory
·
23 May 2025
·
doi:10.1101/2025.05.23.655716
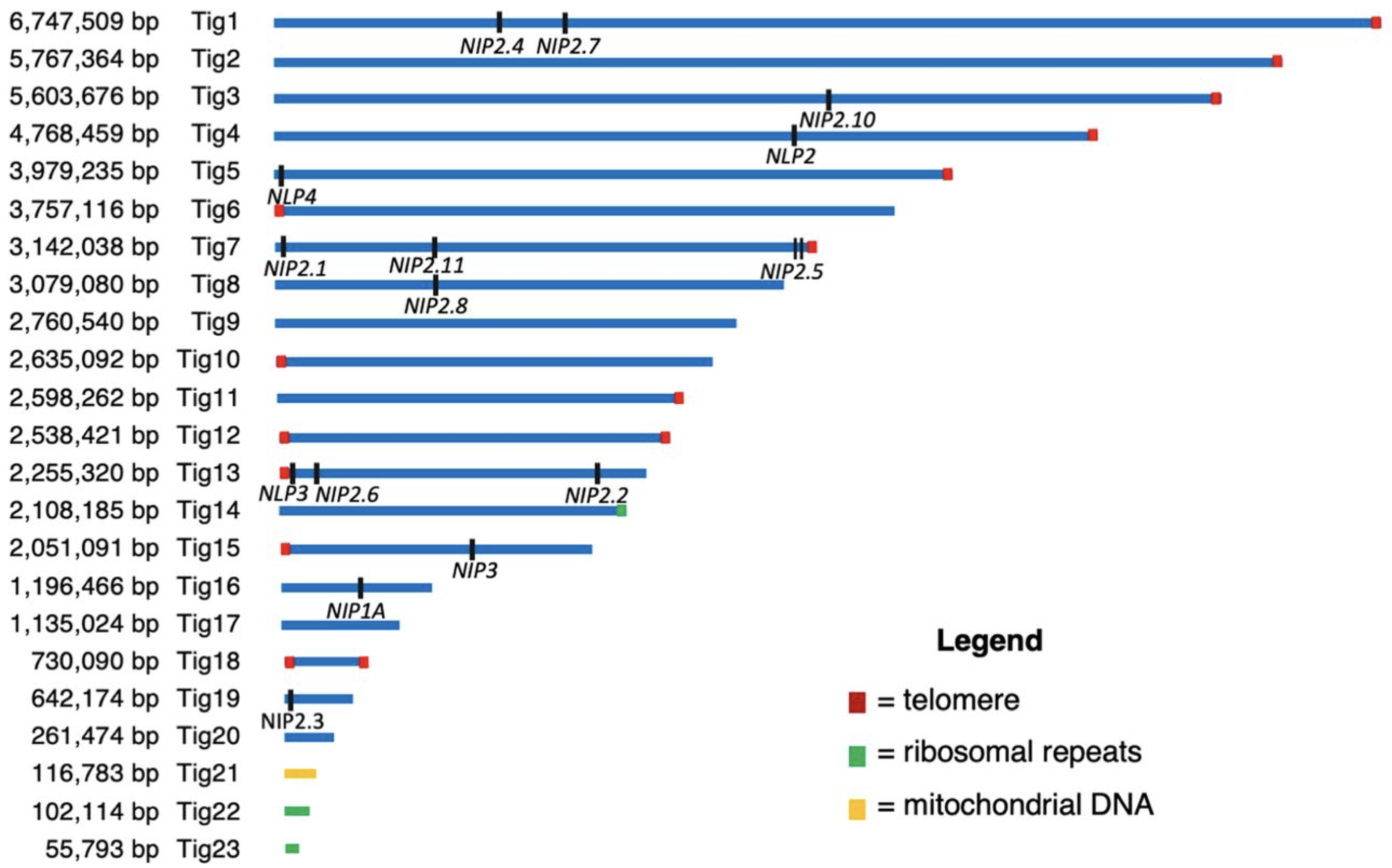
Revisiting the Evolution and Function of NIP2 Paralogues in the Rhynchosporium Spp. Complex
Plant Pathology
·
19 May 2025
·
doi:10.1111/ppa.14111

Corn rust population genomics reveals a cryptic virulent group and adaptive effectorome
Cold Spring Harbor Laboratory
·
12 May 2025
·
doi:10.1101/2025.05.07.652763
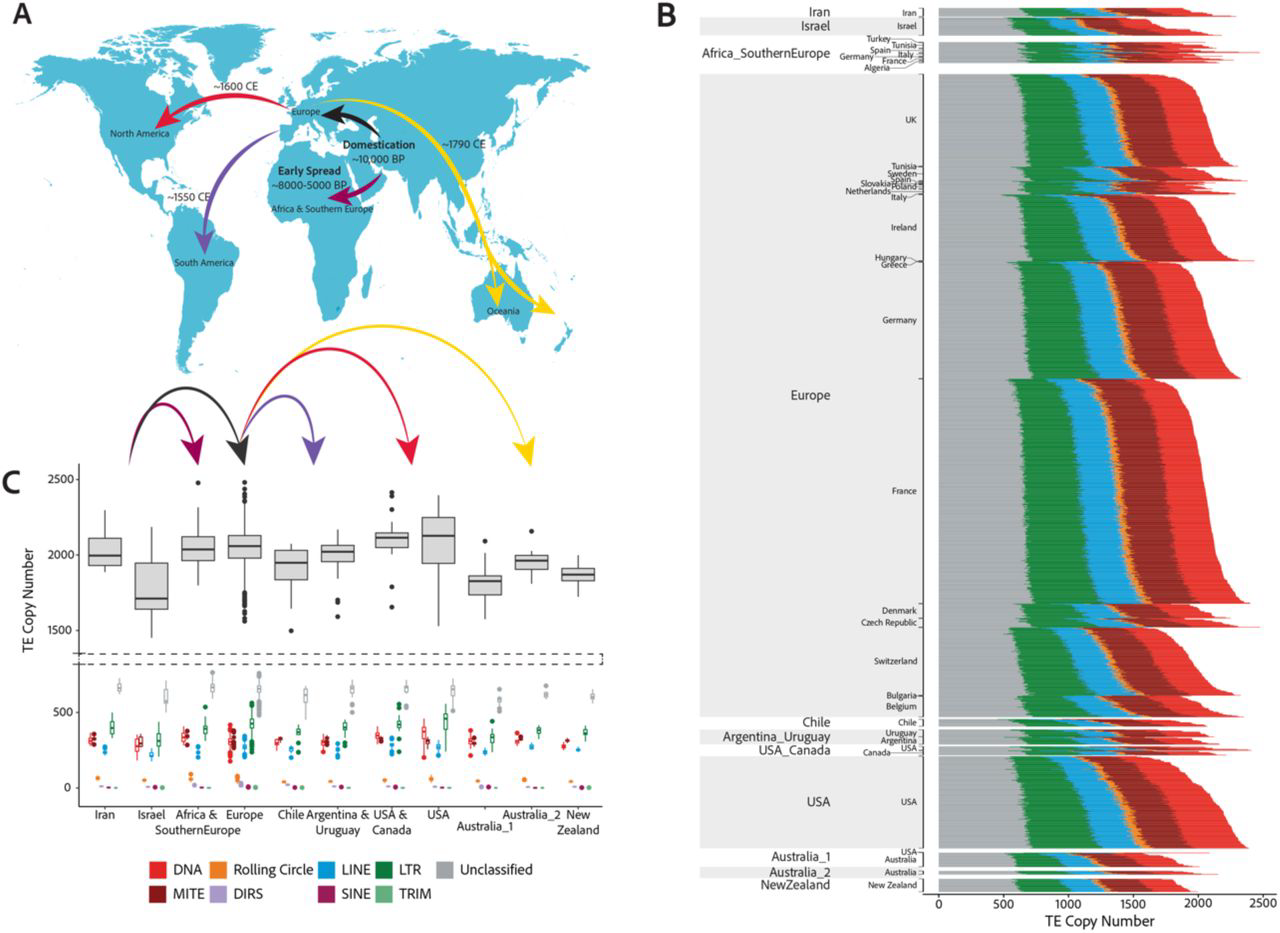
Historic transposon mobilisation waves create distinct pools of adaptive variants in a major crop pathogen
Cold Spring Harbor Laboratory
·
08 Apr 2025
·
doi:10.1101/2025.04.02.646807
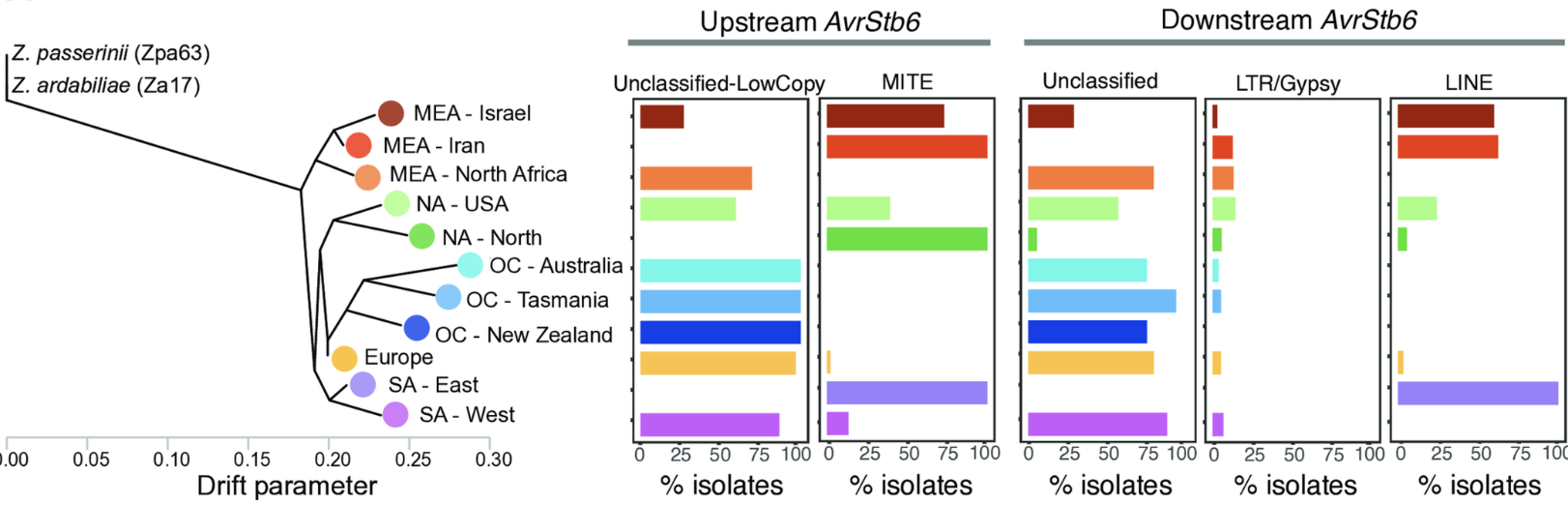
Diversification, loss, and virulence gains of the major effector AvrStb6 during continental spread of the wheat pathogen Zymoseptoria tritici
PLOS Pathogens
·
31 Mar 2025
·
doi:10.1371/journal.ppat.1012983
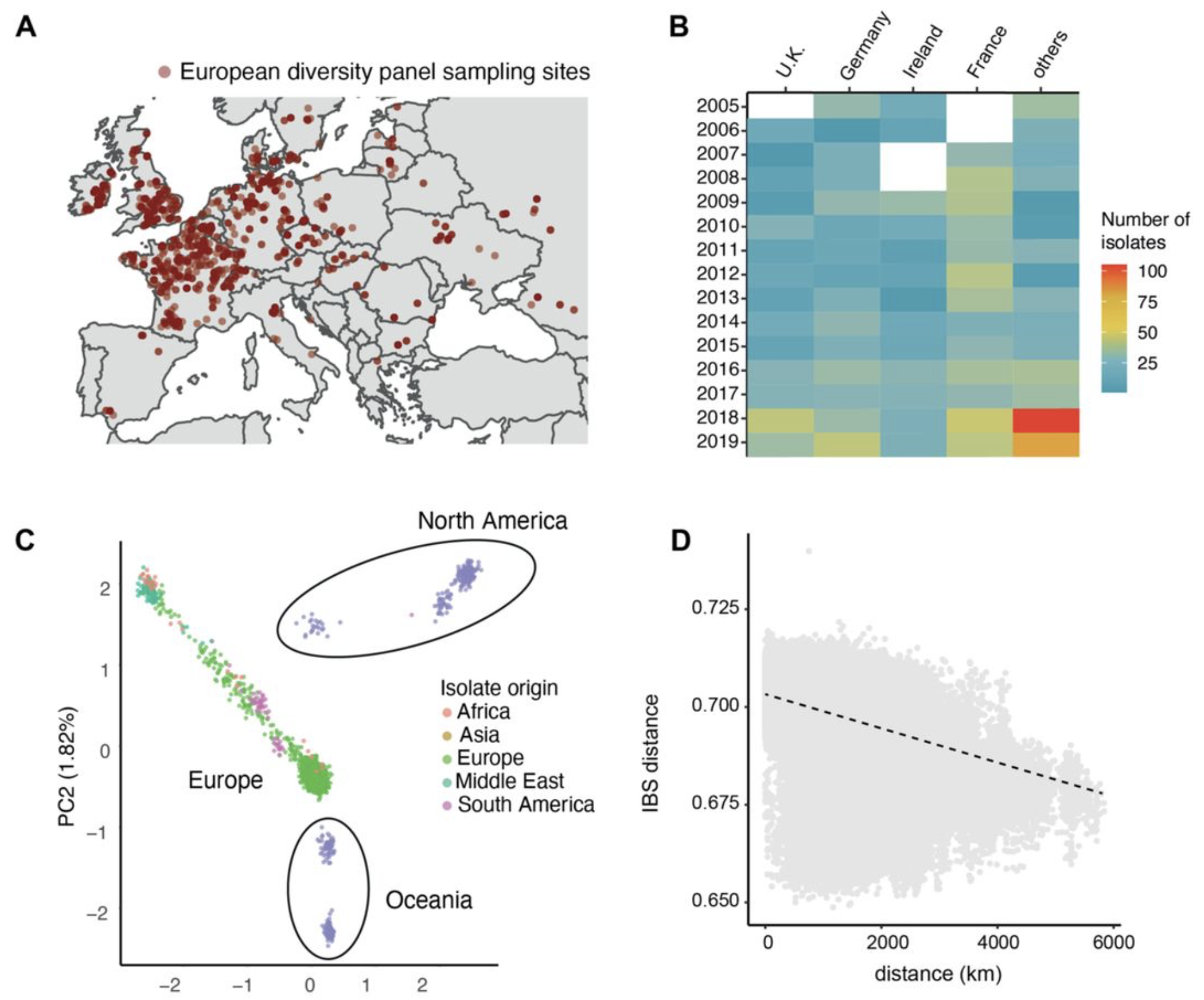
A large European diversity panel reveals complex azole fungicide resistance gains of a major wheat pathogen
Cold Spring Harbor Laboratory
·
19 Mar 2025
·
doi:10.1101/2025.03.19.644200
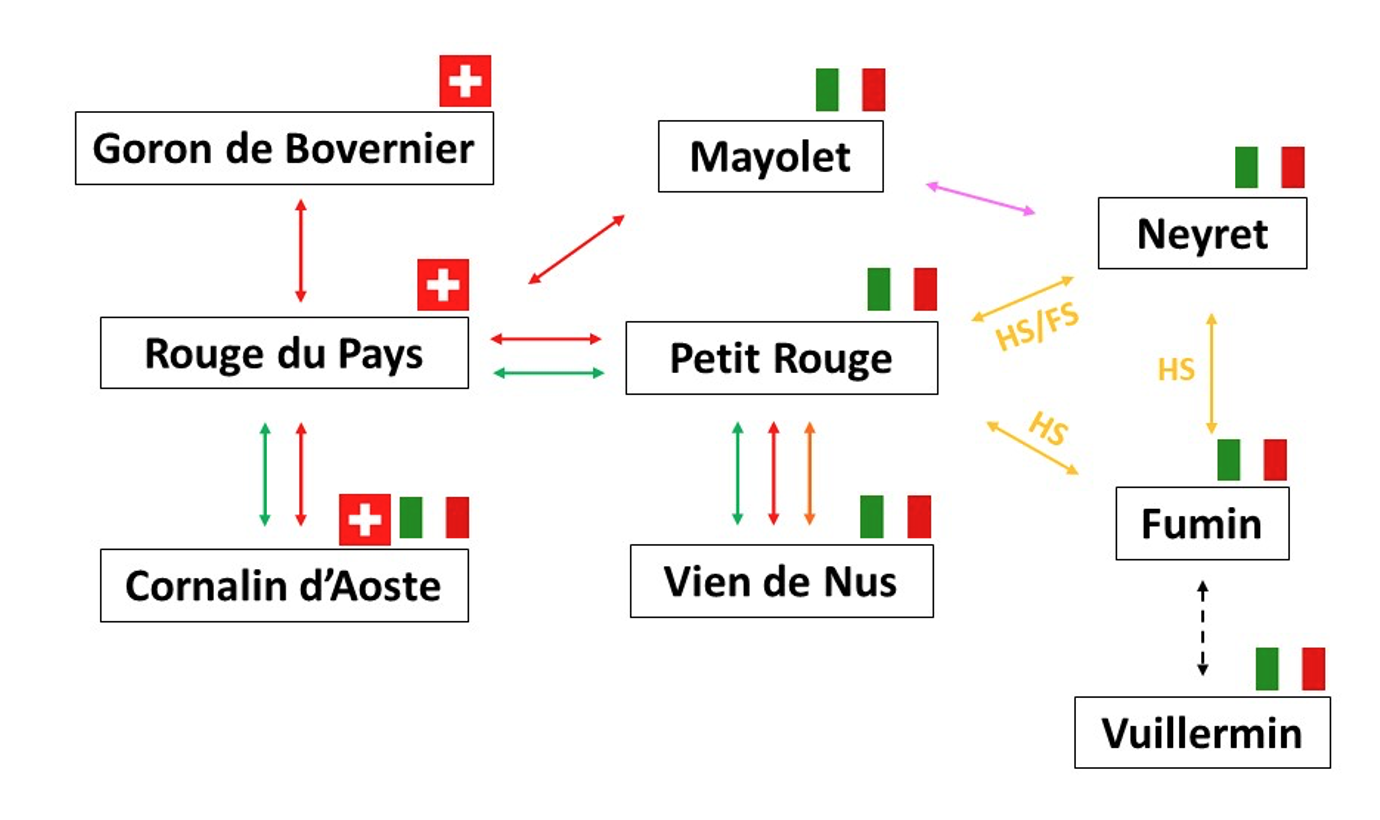
A divergent haplotype with a large deletion at the berry color locus causes a white-skinned phenotype in grapevine
Horticulture Research
·
06 Mar 2025
·
doi:10.1093/hr/uhaf069
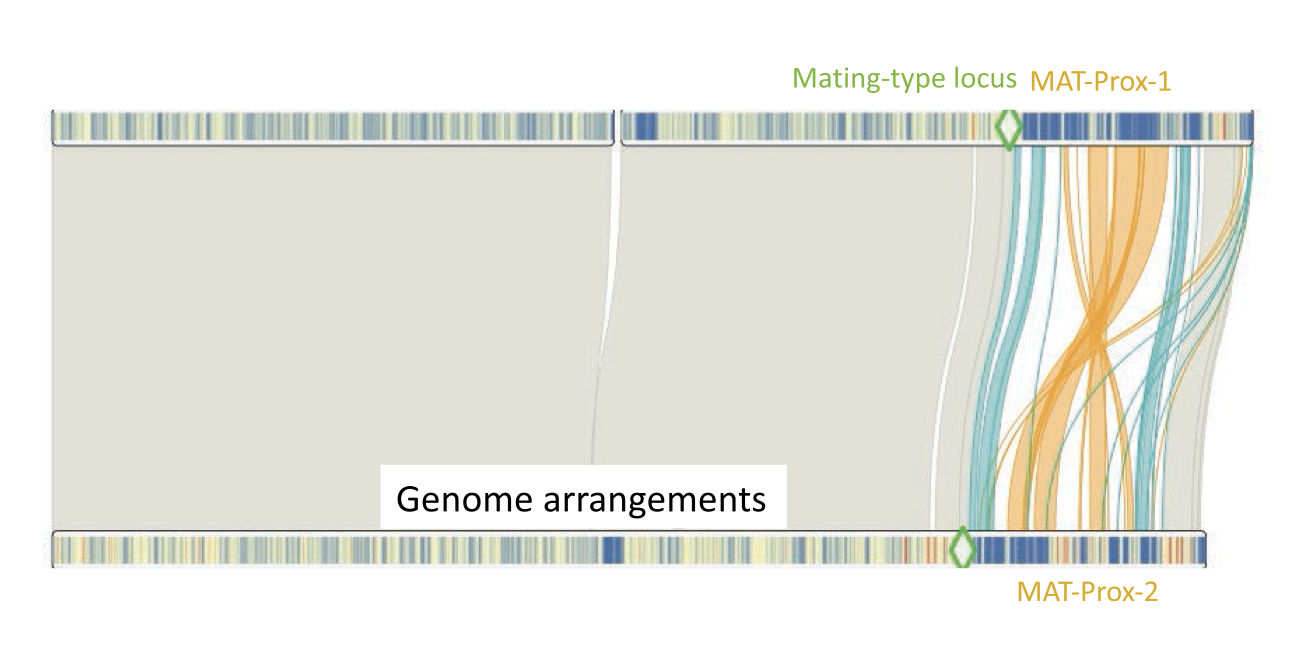
An Inversion Polymorphism Under Balancing Selection, Involving Giant Mobile Elements, in an Invasive Fungal Pathogen
Molecular Biology and Evolution
·
01 Feb 2025
·
doi:10.1093/molbev/msaf026
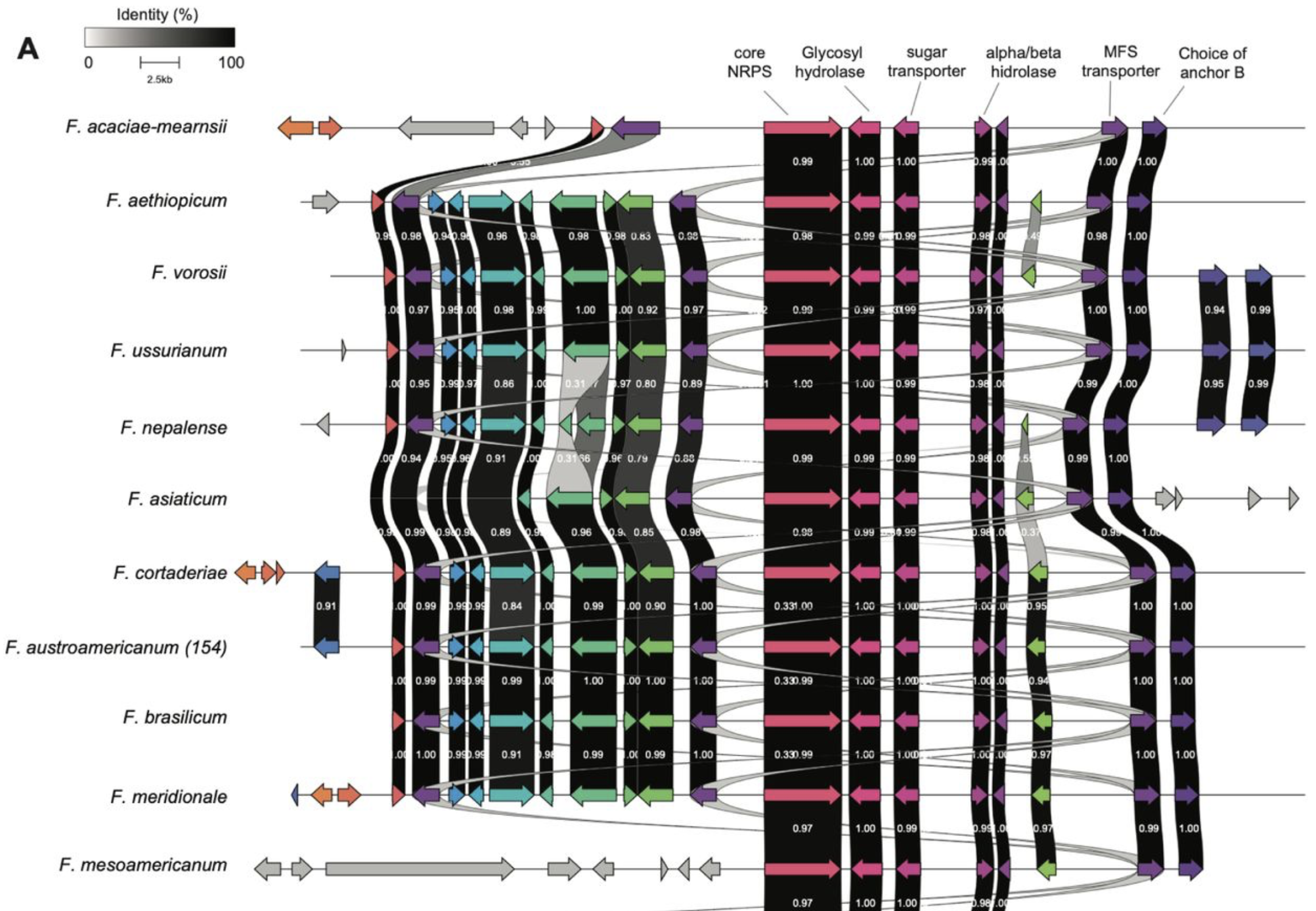
Wheat infection byFusarium graminearumspecies complex members is facilitated by a transcriptionally conserved non-ribosomal peptide synthetase gene cluster
Cold Spring Harbor Laboratory
·
25 Jan 2025
·
doi:10.1101/2025.01.24.634825
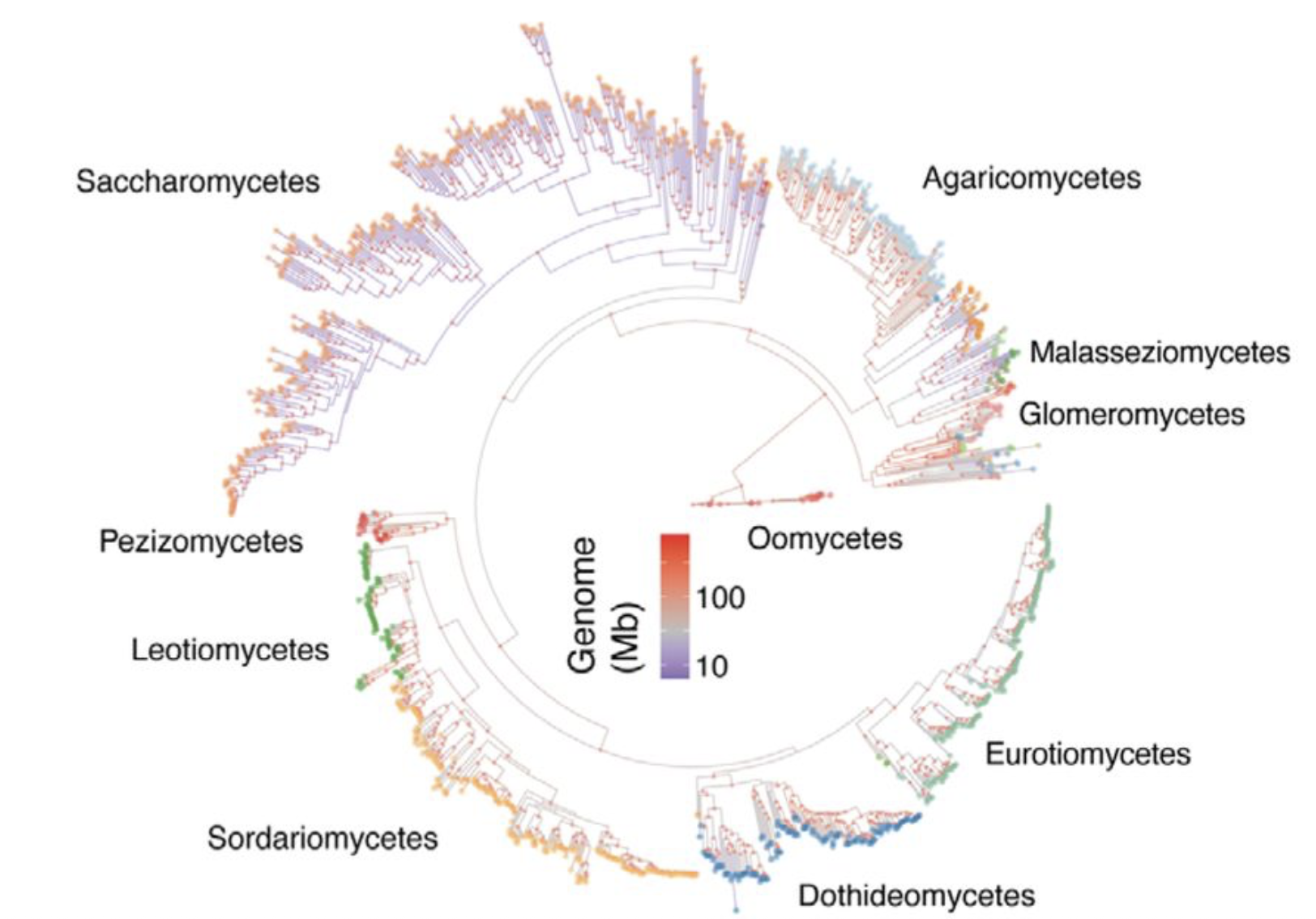
A systematic screen for genetic factors underpinning transposon defense systems across the fungal kingdom
Cold Spring Harbor Laboratory
·
14 Jan 2025
·
doi:10.1101/2025.01.10.632494

Global diversification of the common moonwort ferns (Botrychium lunaria group, Ophioglossaceae) was mainly driven by Pleistocene climatic shifts
Annals of Botany
·
11 Jan 2025
·
doi:10.1093/aob/mcae228

Identifying the pedoclimatic conditions most critical in the susceptibility of a grapevine cultivar to esca disease
OENO One
·
08 Jan 2025
·
doi:10.20870/oeno-one.2025.59.1.7659
2024
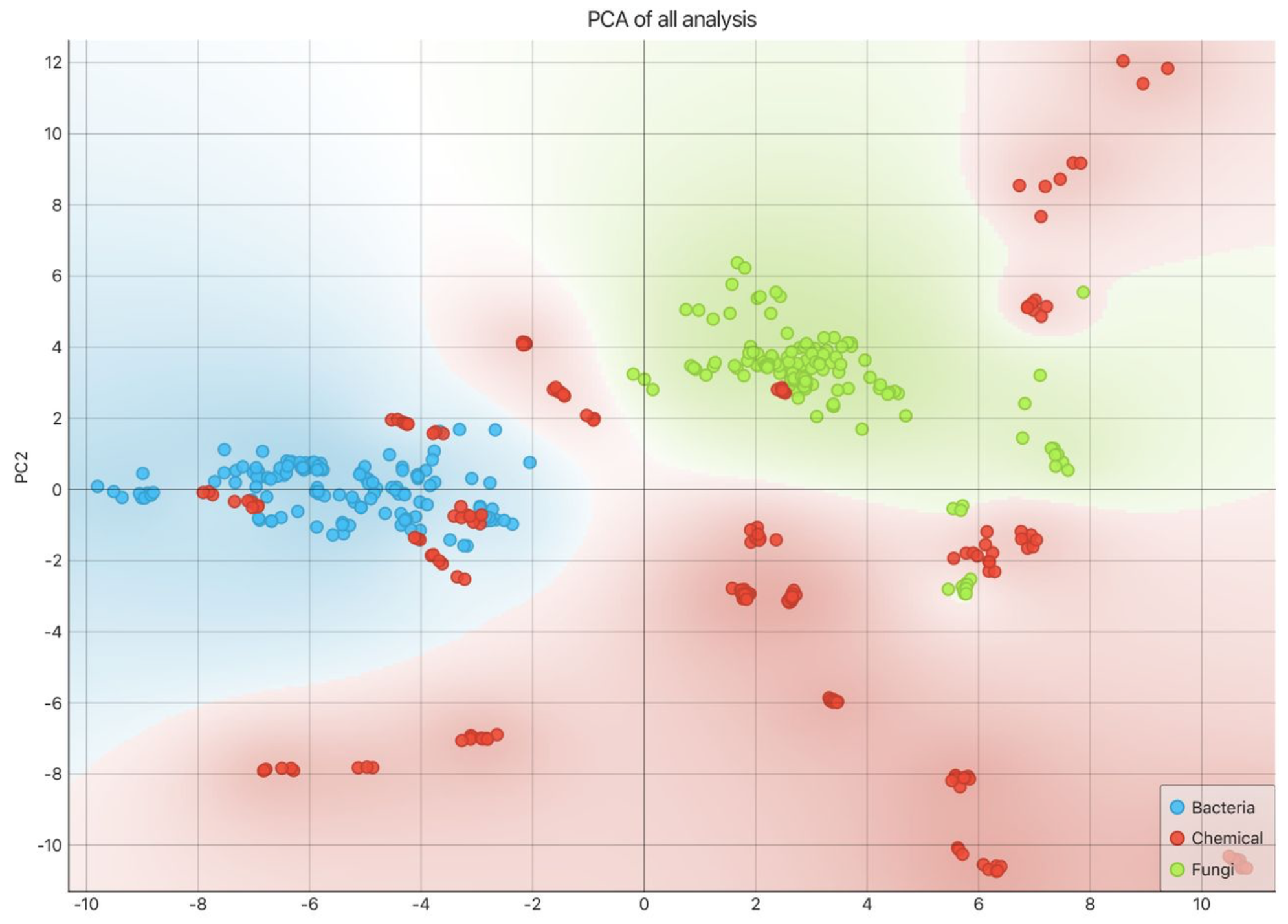
Rapid and In-situ Detection of Biological Agents via Near-Infrared Spectroscopy and Cloud-Powered Neural Networks
Cold Spring Harbor Laboratory
·
15 Nov 2024
·
doi:10.1101/2024.11.15.623740
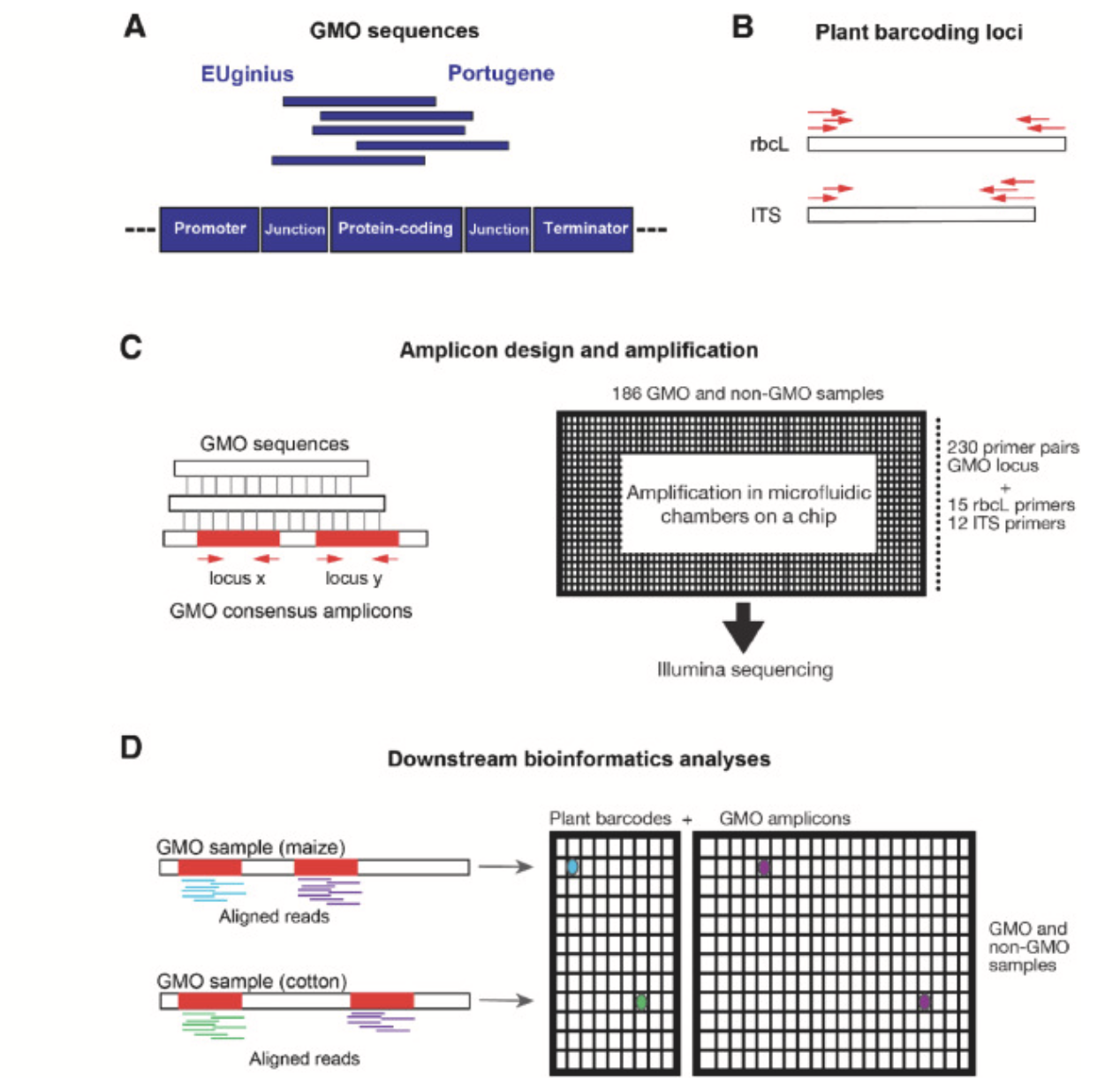
Detection of genetically modified organisms using highly multiplexed amplicon sequencing
Food Control
·
01 Nov 2024
·
doi:10.1016/j.foodcont.2024.110670
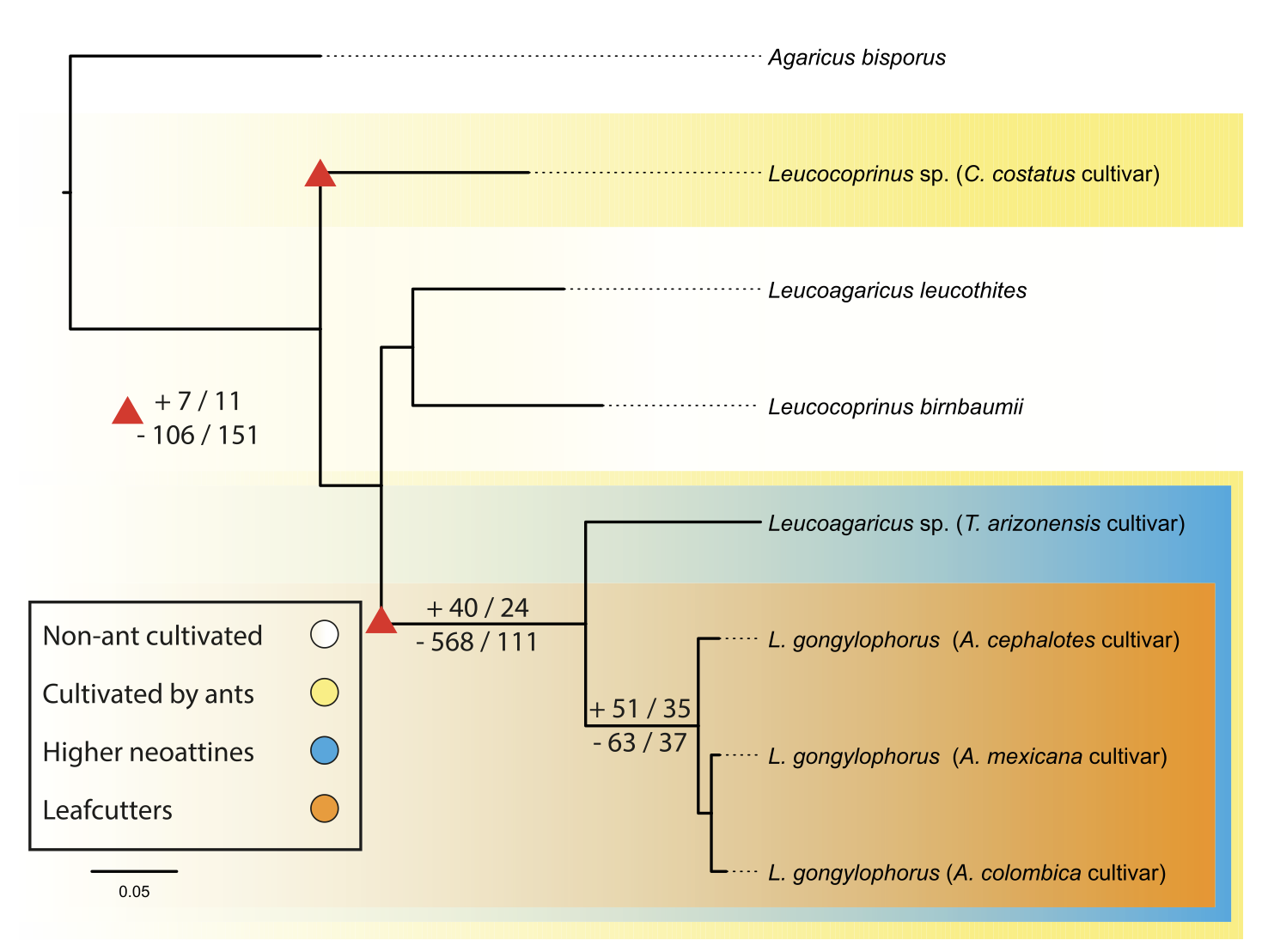
Genomic Signatures of Domestication in a Fungus Obligately Farmed by Leafcutter Ants
Molecular Biology and Evolution
·
17 Sep 2024
·
doi:10.1093/molbev/msae197
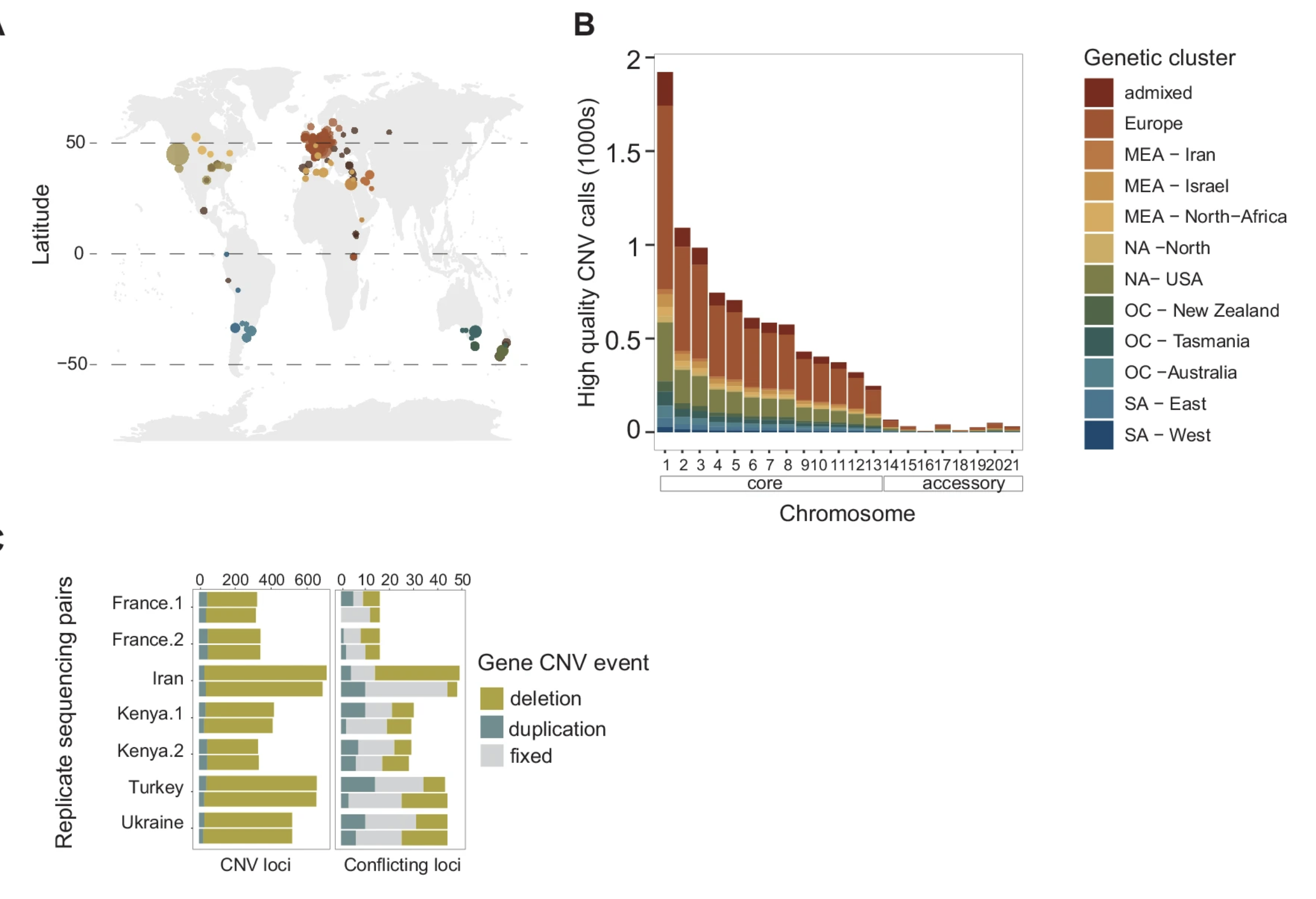
Copy number variation introduced by a massive mobile element facilitates global thermal adaptation in a fungal wheat pathogen
Nature Communications
·
08 Jul 2024
·
doi:10.1038/s41467-024-49913-7
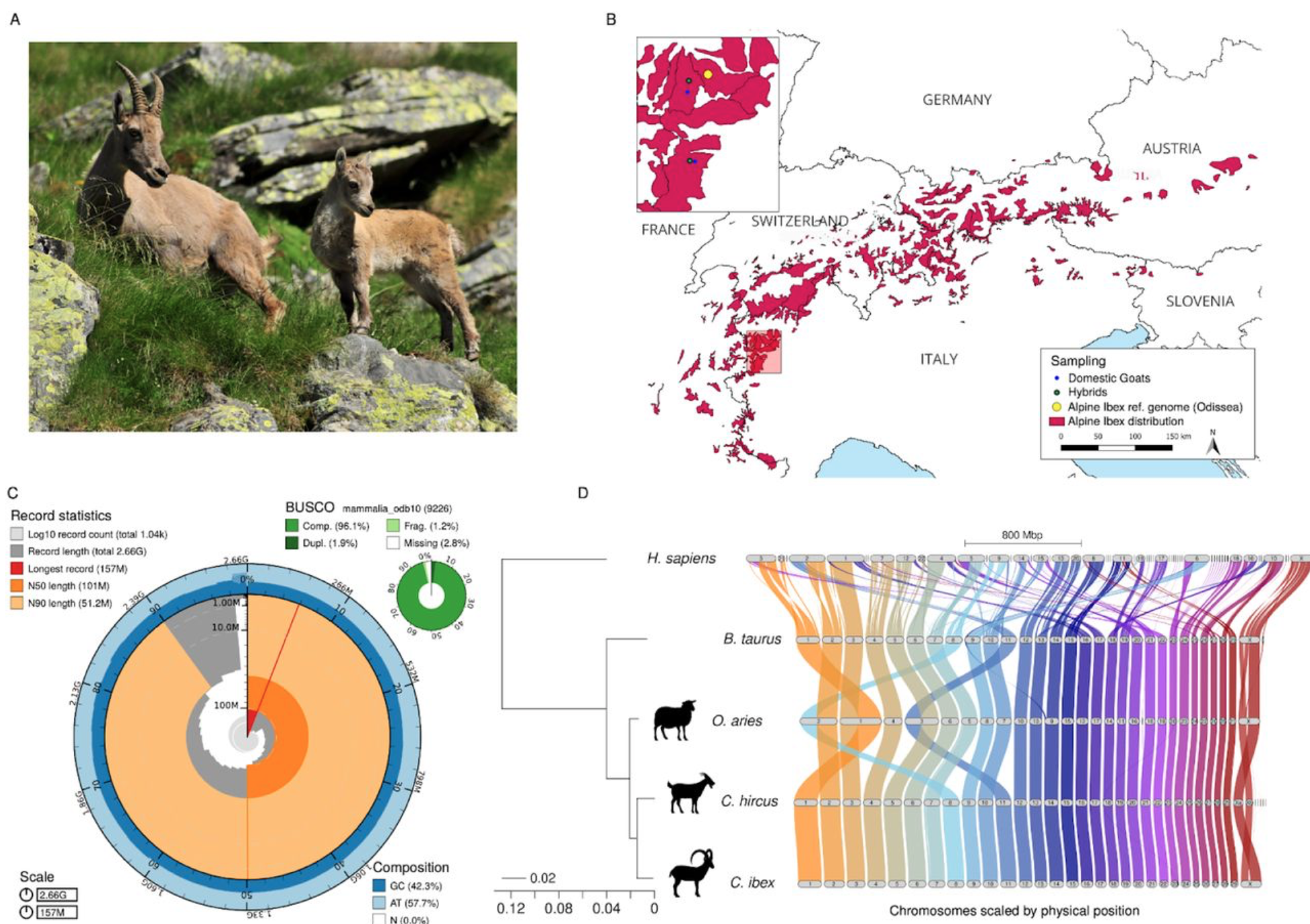
Hybridization load introduced by Alpine ibex hybrid swarms
Cold Spring Harbor Laboratory
·
27 Jun 2024
·
doi:10.1101/2024.06.24.599926
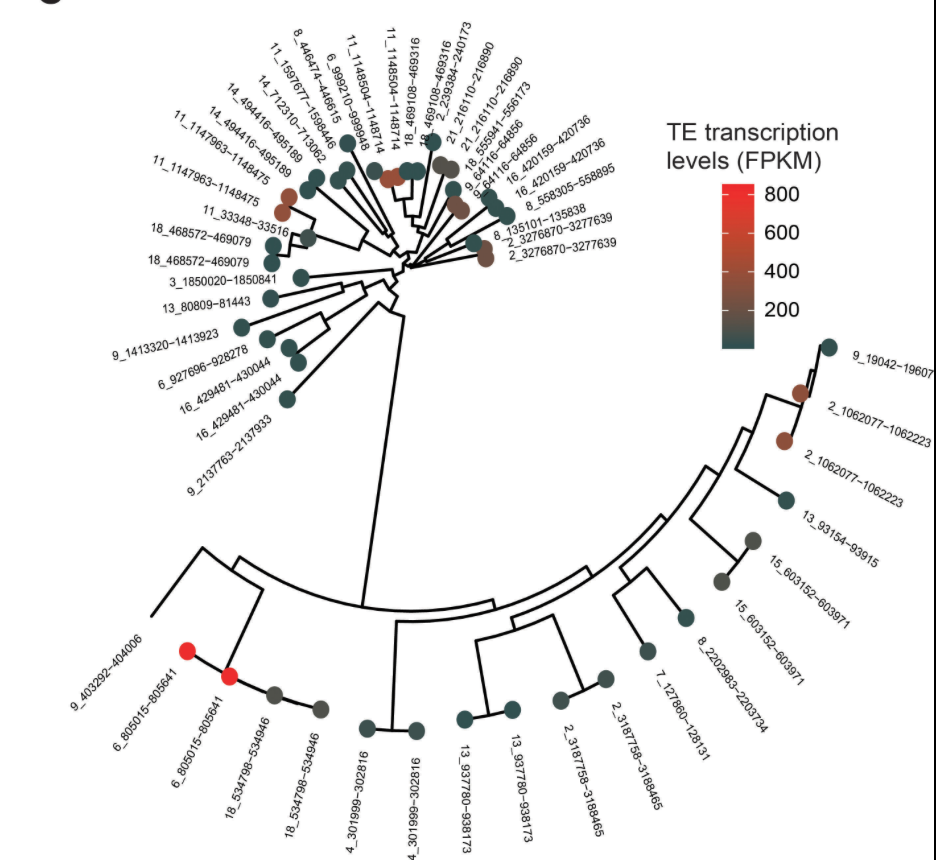
Population-level transposable element expression dynamics influence trait evolution in a fungal crop pathogen
mBio
·
13 Mar 2024
·
doi:10.1128/mbio.02840-23
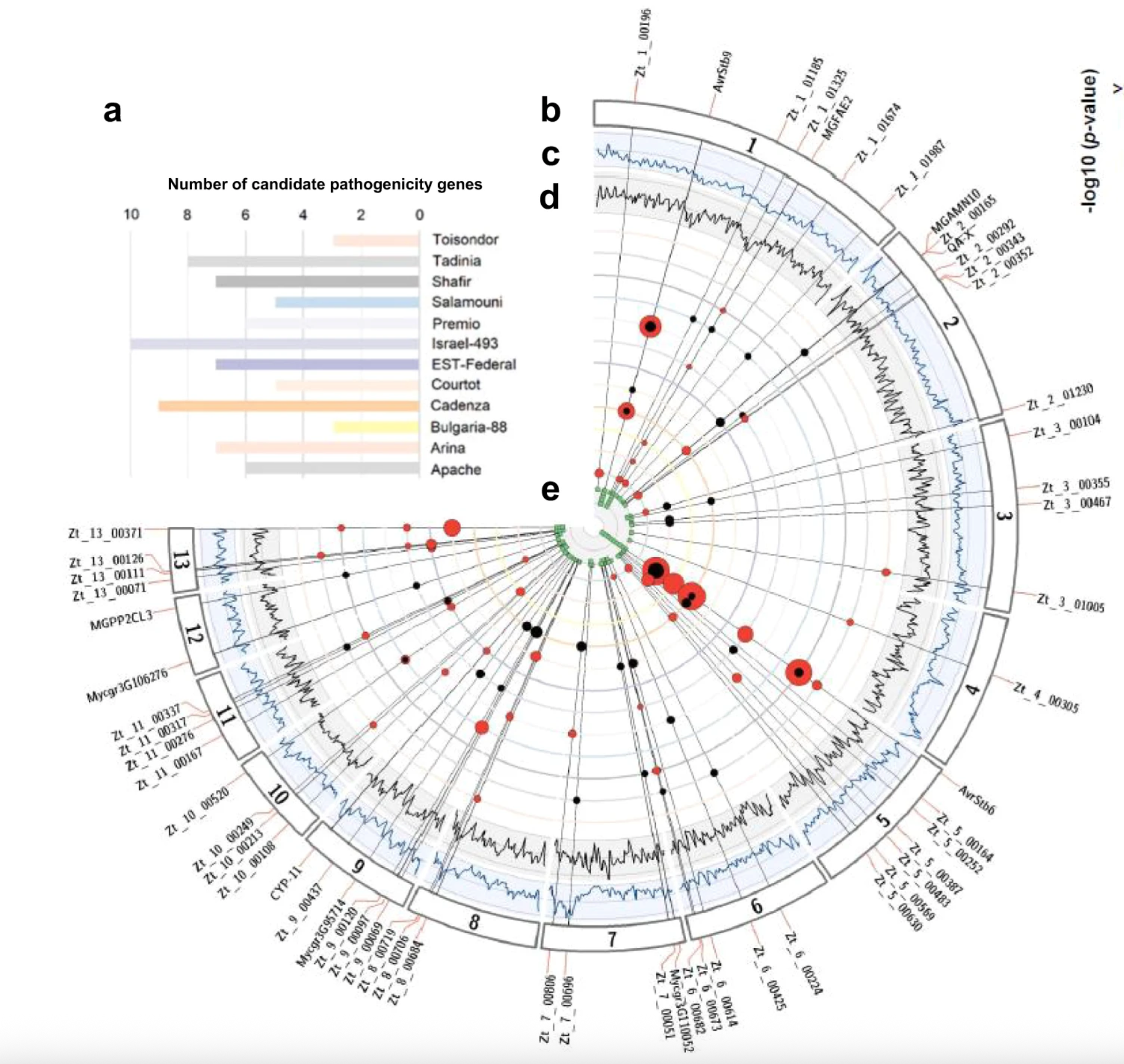
Quantitative pathogenicity and host adaptation in a fungal plant pathogen revealed by whole-genome sequencing
Nature Communications
·
02 Mar 2024
·
doi:10.1038/s41467-024-46191-1

Dimensions of genome dynamics in fungal pathogens: from fundamentals to applications
BMC Biology
·
26 Jan 2024
·
doi:10.1186/s12915-023-01786-w
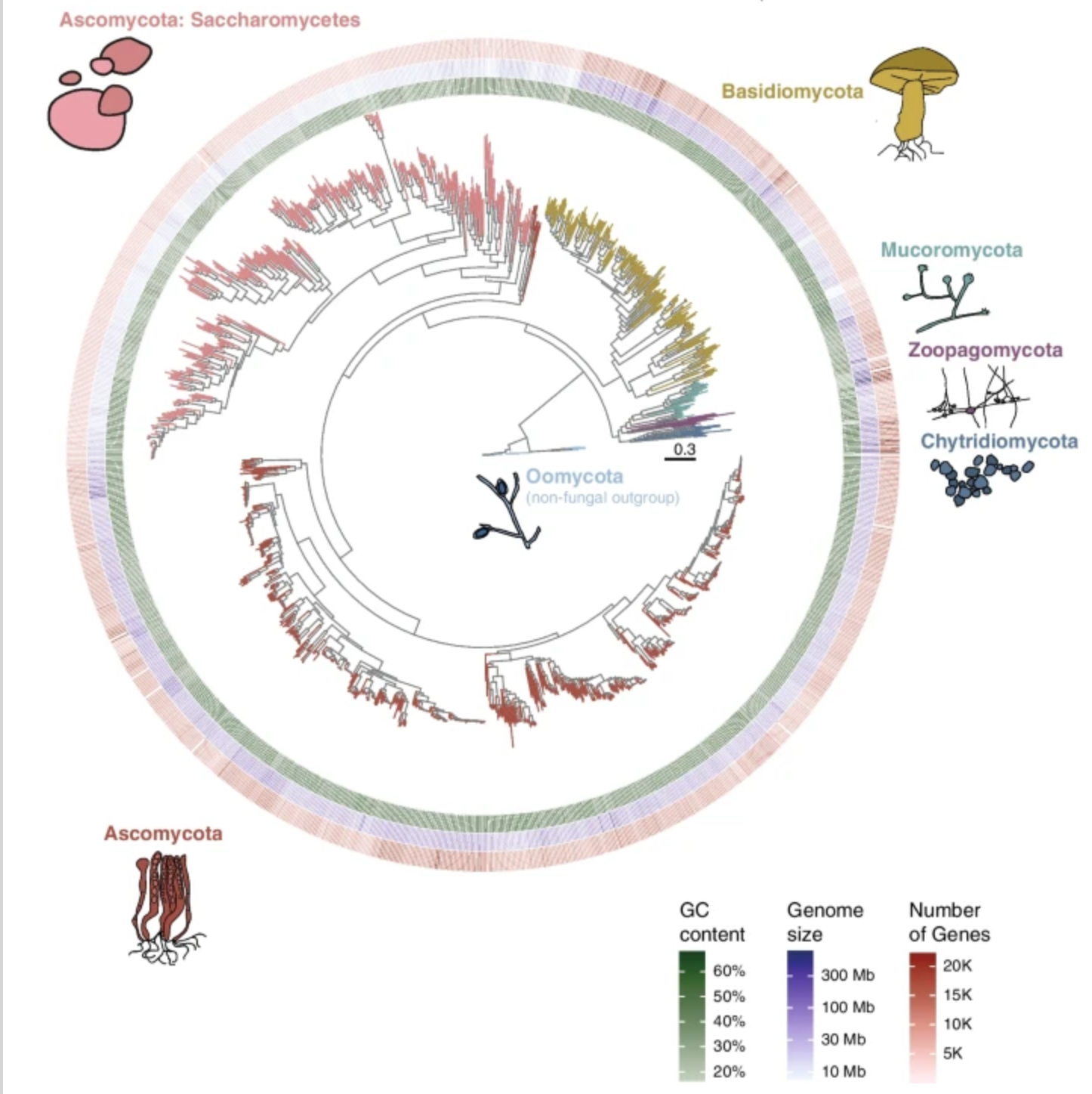
A systematic screen for co-option of transposable elements across the fungal kingdom
Mobile DNA
·
20 Jan 2024
·
doi:10.1186/s13100-024-00312-1

Le Blanchier: redécouverte d’un ancien cépage dans le vignoble valaisan
Agroscope
·
01 Jan 2024
·
doi:10.34776/afs15-207
2023
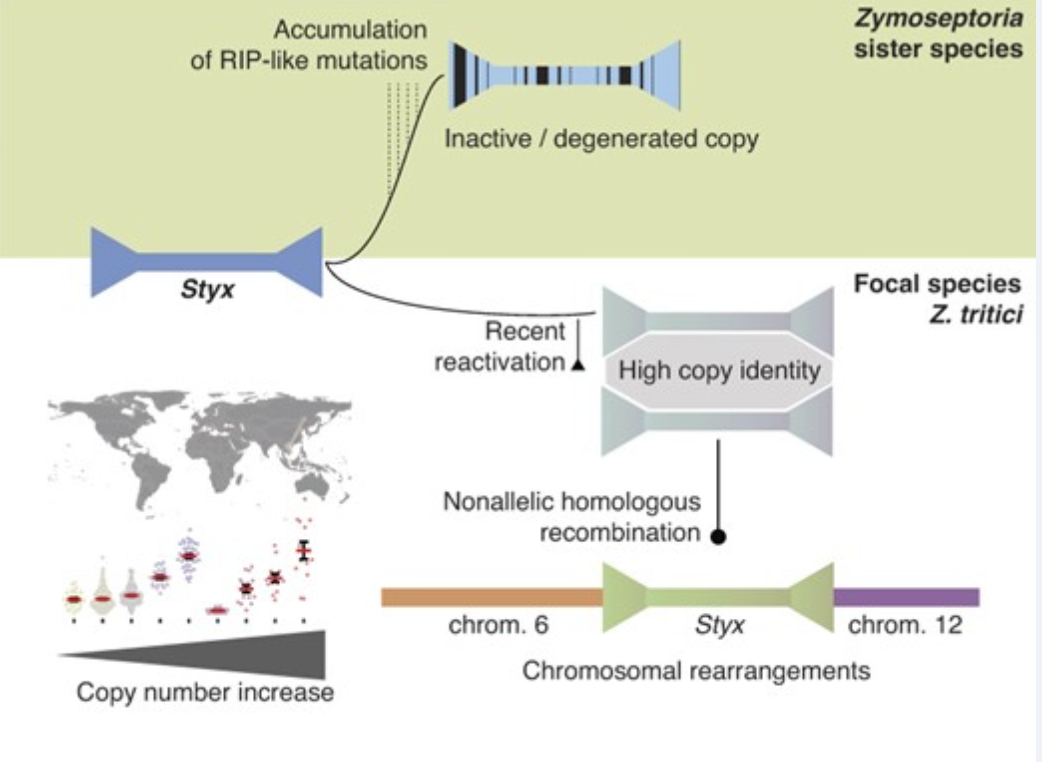
Recent reactivation of a pathogenicity-associated transposable element is associated with major chromosomal rearrangements in a fungal wheat pathogen
Nucleic Acids Research
·
24 Dec 2023
·
doi:10.1093/nar/gkad1214

Two‐speed genomes of Epichloe fungal pathogens show contrasting signatures of selection between species and across populations
Molecular Ecology
·
12 Dec 2023
·
doi:10.1111/mec.17242

The expression landscape and pangenome of long non-coding RNA in the fungal wheat pathogen Zymoseptoria tritici
Microbial Genomics
·
22 Nov 2023
·
doi:10.1099/mgen.0.001136
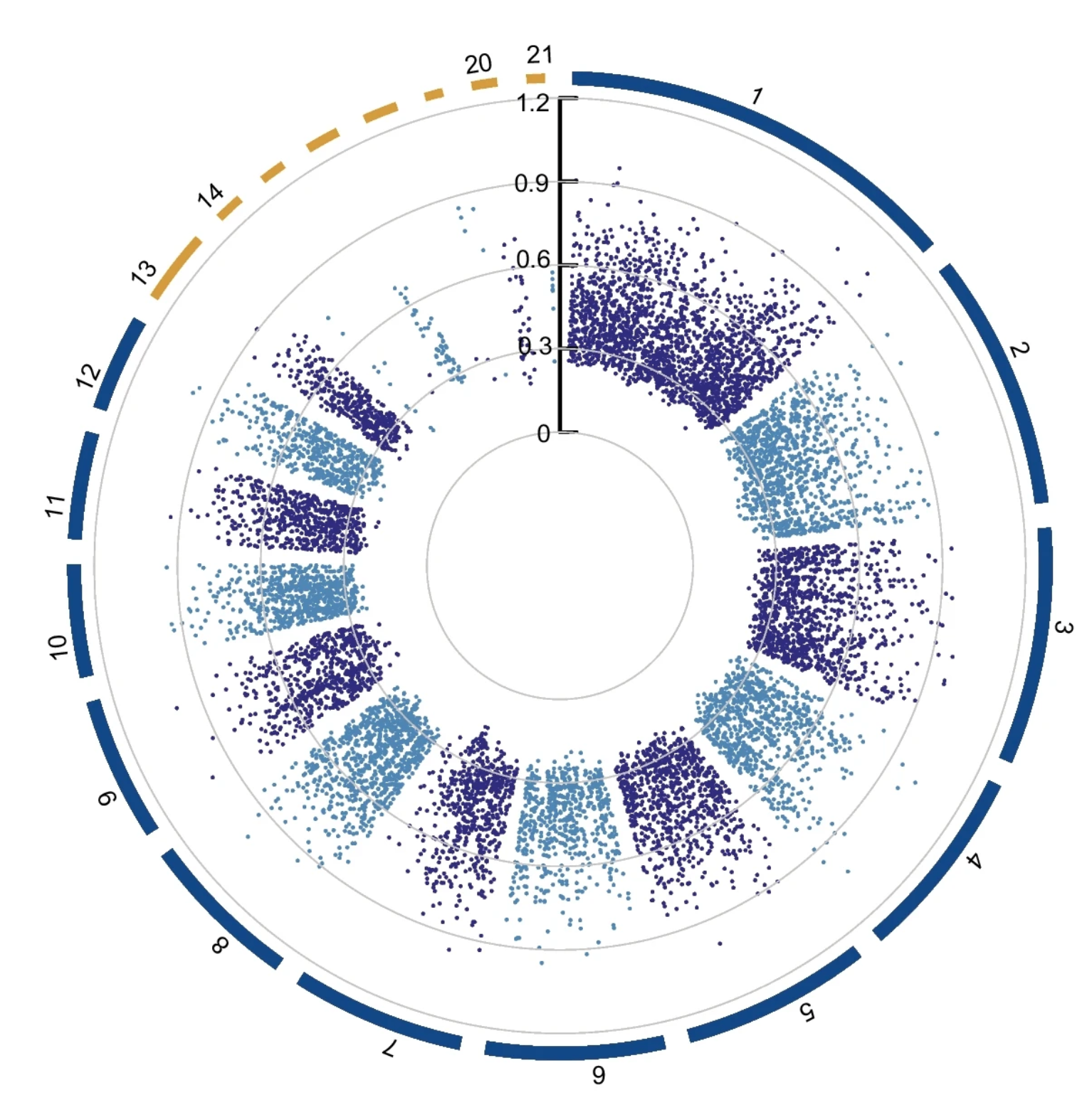
Genome-wide expression QTL mapping reveals the highly dynamic regulatory landscape of a major wheat pathogen
BMC Biology
·
20 Nov 2023
·
doi:10.1186/s12915-023-01763-3

A pangenome-guided manually curated library of transposable elements for Zymoseptoria tritici
BMC Research Notes
·
16 Nov 2023
·
doi:10.1186/s13104-023-06613-7
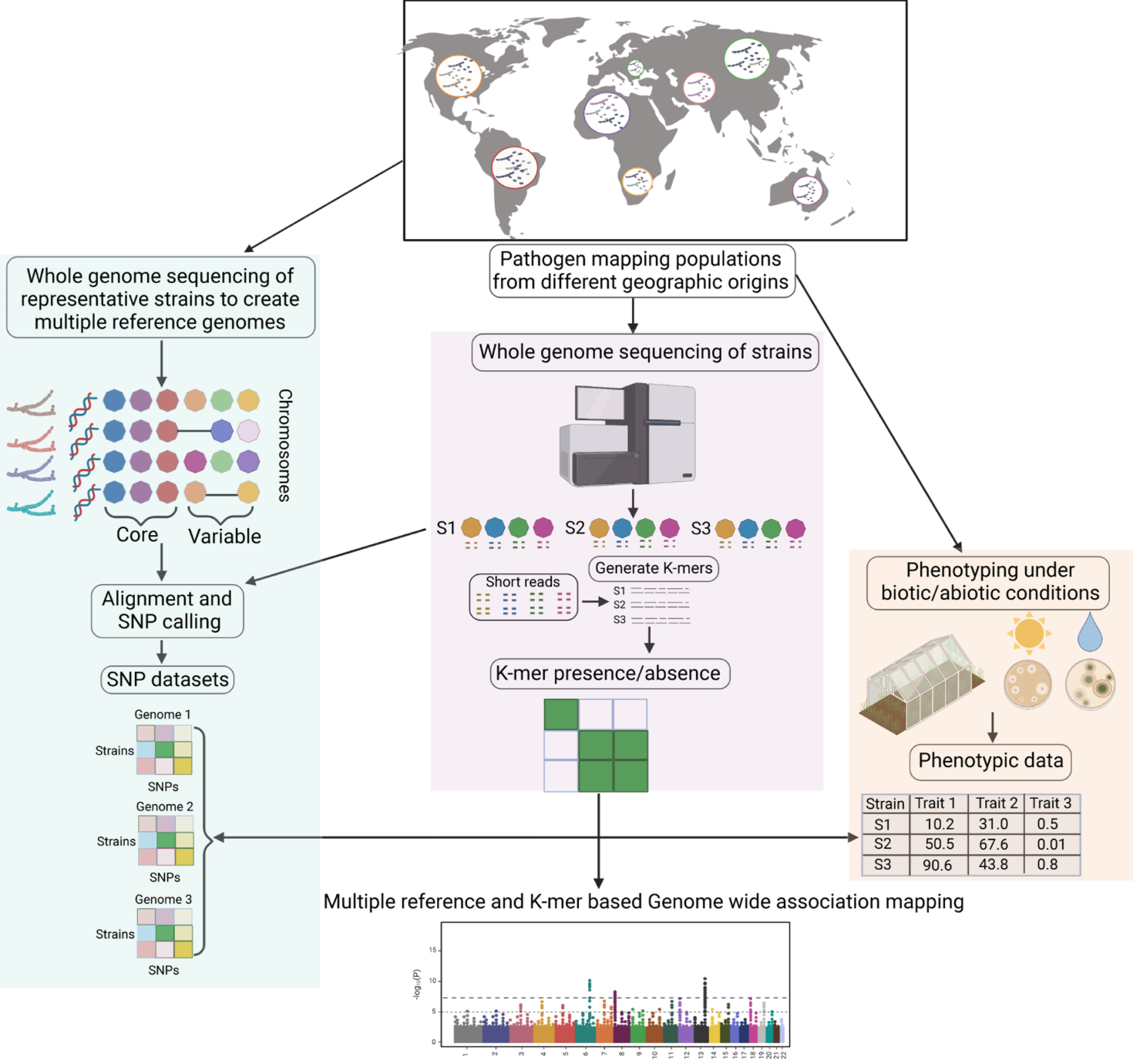
Combined reference-free and multi-reference based GWAS uncover cryptic variation underlying rapid adaptation in a fungal plant pathogen
PLOS Pathogens
·
16 Nov 2023
·
10.1371/journal.ppat.1011801

Virulence Associations and Global Context of AvrStb6 Genetic Diversity in Iranian Populations of Zymoseptoria tritici
Phytopathology®
·
10 Nov 2023
·
doi:10.1094/PHYTO-09-22-0348-R
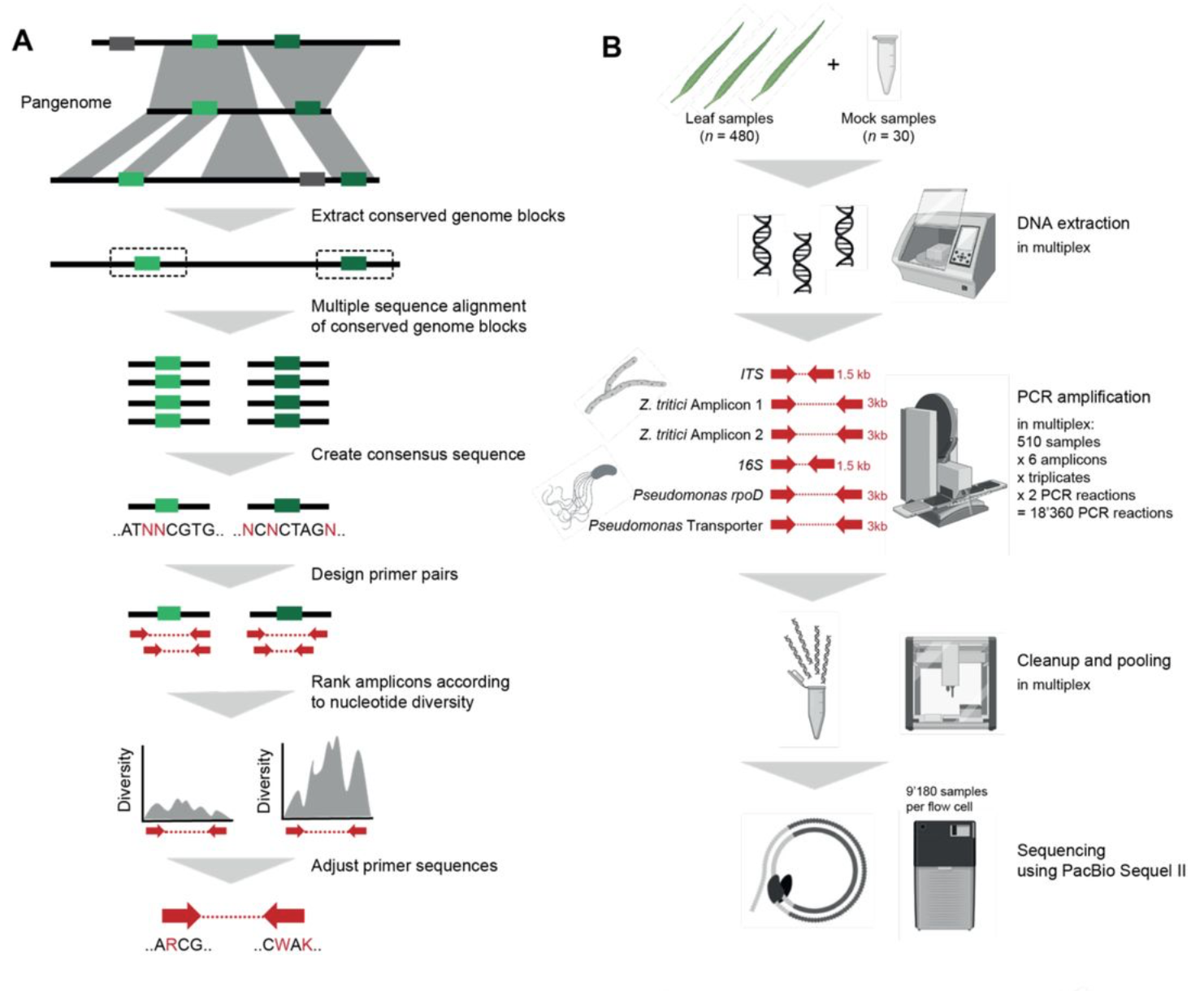
High-resolution profiling of bacterial and fungal communities using pangenome-informed taxon-specific long-read amplicons
Cold Spring Harbor Laboratory
·
17 Jul 2023
·
doi:10.1101/2023.07.17.549274
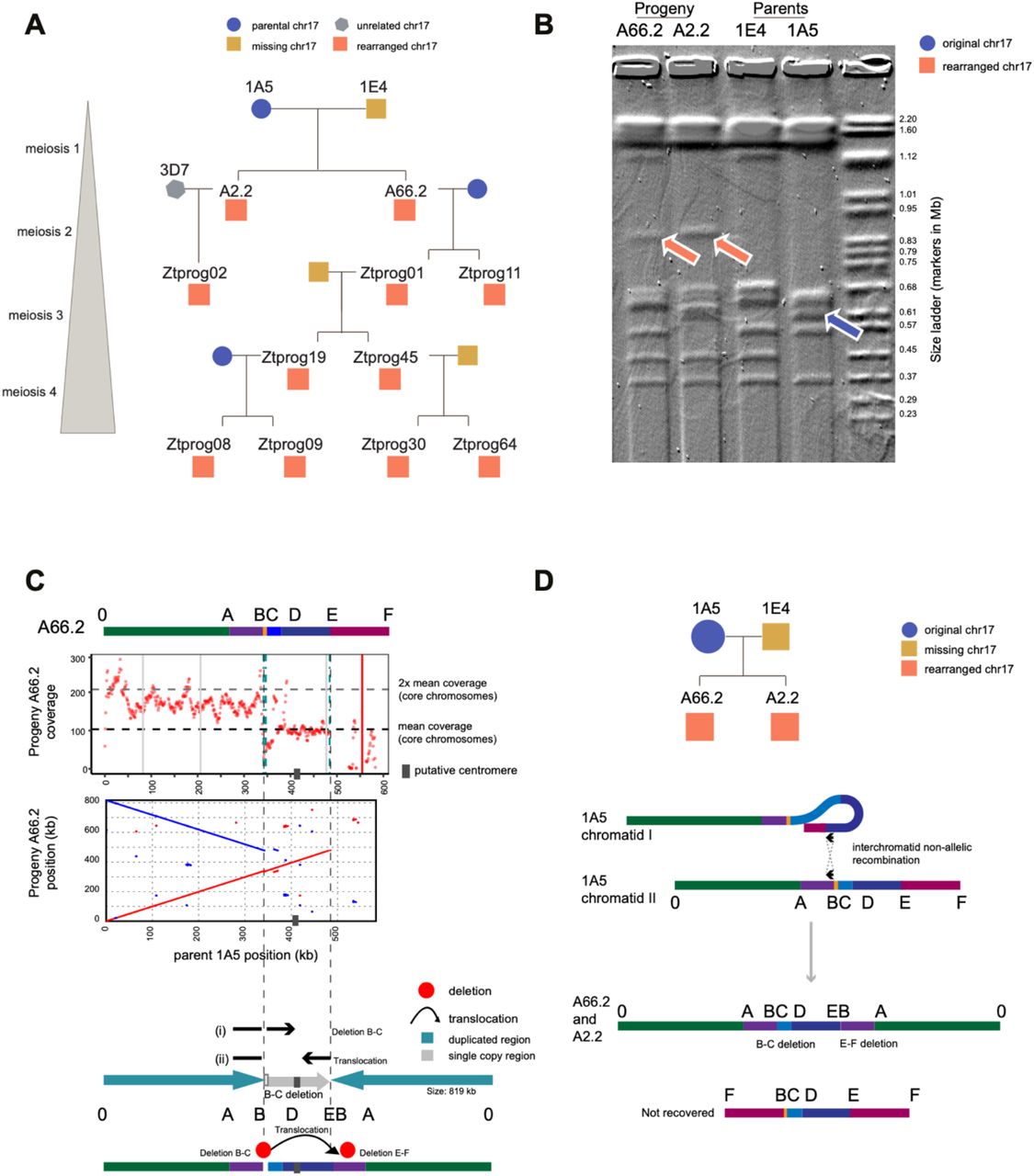
Recurrent chromosome destabilization through repeat-mediated rearrangements in a fungal pathogen
Cold Spring Harbor Laboratory
·
15 Jul 2023
·
doi:10.1101/2023.07.14.549097

How genomics can help biodiversity conservation
Trends in Genetics
·
01 Jul 2023
·
doi:10.1016/j.tig.2023.01.005
A secreted protease-like protein in Zymoseptoria tritici is responsible for avirulence on Stb9 resistance gene in wheat
PLOS Pathogens
·
12 May 2023
·
doi:10.1371/journal.ppat.1011376
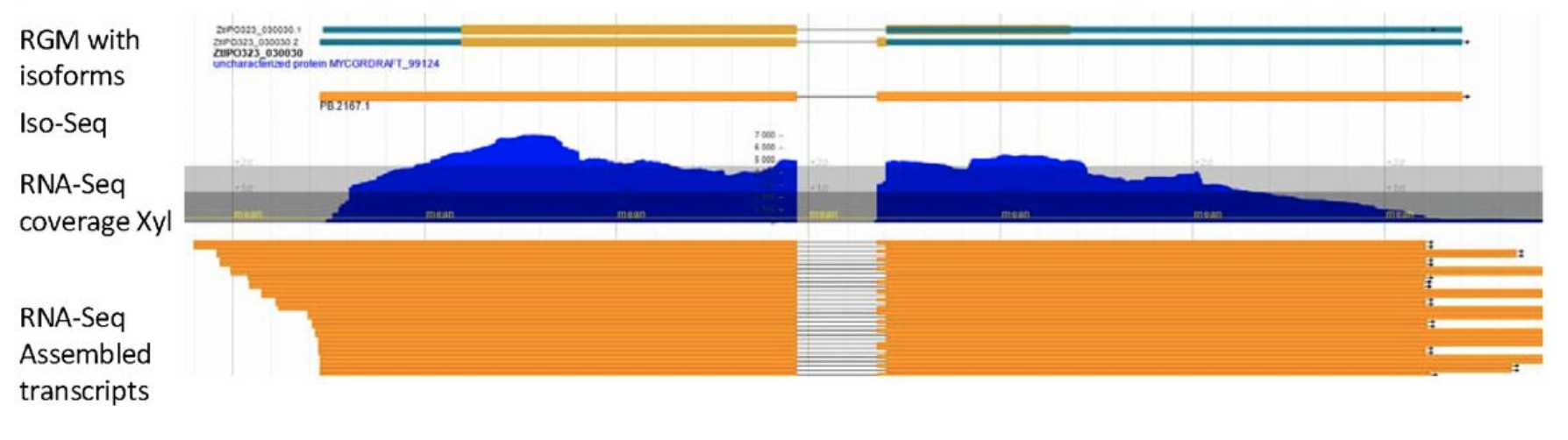
Improved gene annotation of the fungal wheat pathogenZymoseptoria triticibased on combined Iso-Seq and RNA-Seq evidence
Cold Spring Harbor Laboratory
·
28 Apr 2023
·
doi:10.1101/2023.04.26.537486

Divergent Outcomes of Direct Conspecific Pathogen Strain Interaction and Plant Co-Infection Suggest Consequences for Disease Dynamics
Microbiology Spectrum
·
13 Apr 2023
·
doi:10.1128/spectrum.04443-22
Genomic surveillance uncovers a pandemic clonal lineage of the wheat blast fungus
PLOS Biology
·
11 Apr 2023
·
doi:10.1371/journal.pbio.3002052
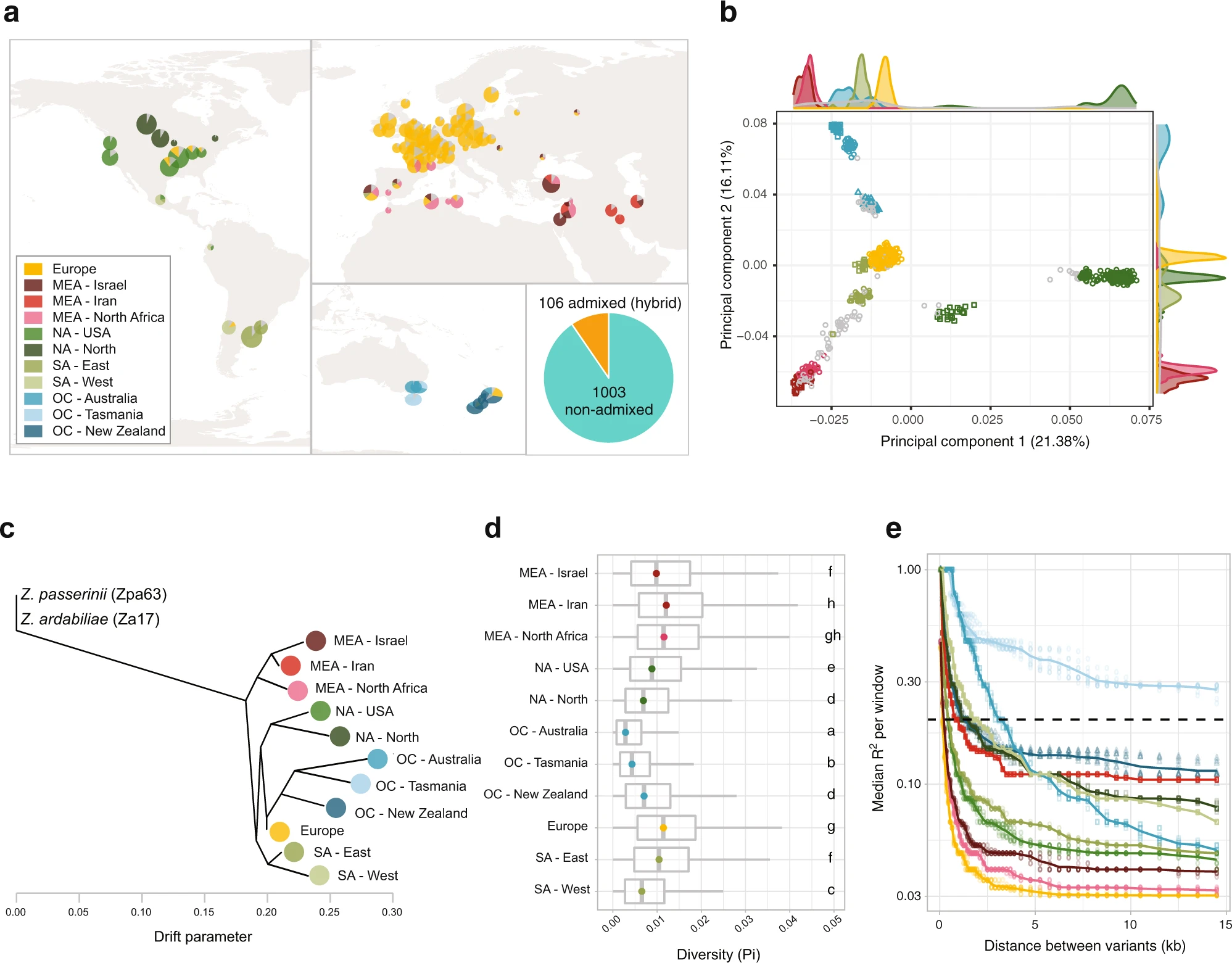
A thousand-genome panel retraces the global spread and adaptation of a major fungal crop pathogen
Nature Communications
·
24 Feb 2023
·
doi:10.1038/s41467-023-36674-y

Recent transposable element bursts are associated with the proximity to genes in a fungal plant pathogen
PLOS Pathogens
·
14 Feb 2023
·
doi:10.1371/journal.ppat.1011130
A highly multiplexed assay to monitor pathogenicity, fungicide resistance and gene flow in the fungal wheat pathogen Zymoseptoria tritici
PLOS ONE
·
06 Feb 2023
·
doi:10.1371/journal.pone.0281181

Genome Evolution in Fungal Plant Pathogens: From Populations to Kingdom-Wide Dynamics
The Mycota
·
01 Jan 2023
·
doi:10.1007/978-3-031-29199-9_5
2022

Quantifying Trade-Offs in the Choice of Ribosomal Barcoding Markers for Fungal Amplicon Sequencing: a Case Study on the Grapevine Trunk Mycobiome
Microbiology Spectrum
·
21 Dec 2022
·
doi:10.1128/spectrum.02513-22
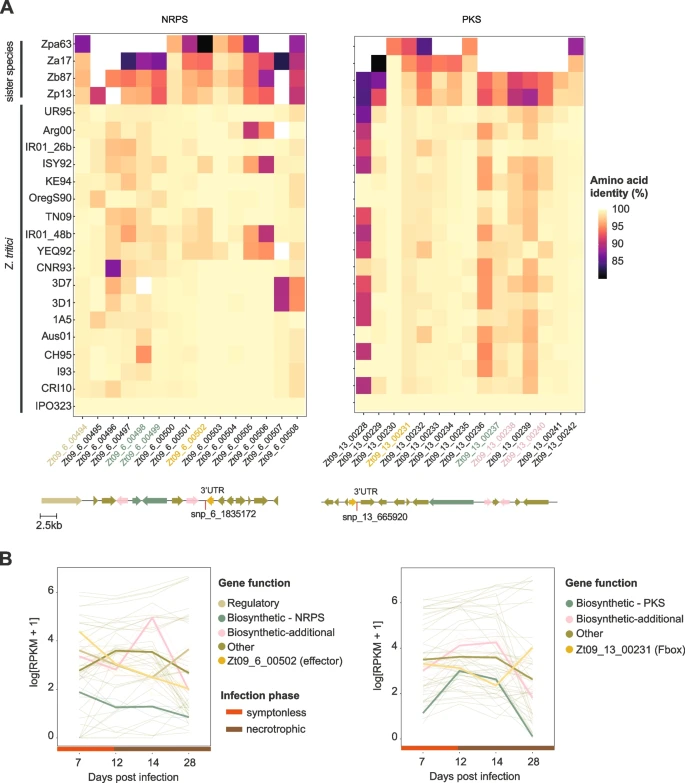
Genome-wide association mapping reveals genes underlying population-level metabolome diversity in a fungal crop pathogen
BMC Biology
·
08 Oct 2022
·
doi:10.1186/s12915-022-01422-z

Unraveling coevolutionary dynamics using ecological genomics
Trends in Genetics
·
01 Oct 2022
·
doi:10.1016/j.tig.2022.05.008

Genome of Malassezia arunalokei and Its Distribution on Facial Skin
Microbiology Spectrum
·
29 Jun 2022
·
doi:10.1128/spectrum.00506-22

Organic acids and glucose prime late-stage fungal biotrophy in maize
Science
·
10 Jun 2022
·
doi:10.1126/science.abo2401

The population genetics of adaptation through copy number variation in a fungal plant pathogen
Molecular Ecology
·
05 Apr 2022
·
doi:10.1111/mec.16435

The era of reference genomes in conservation genomics
Trends in Ecology & Evolution
·
01 Mar 2022
·
doi:10.1016/j.tree.2021.11.008

A devil's bargain with transposable elements in plant pathogens
Trends in Genetics
·
01 Mar 2022
·
doi:10.1016/j.tig.2021.08.005
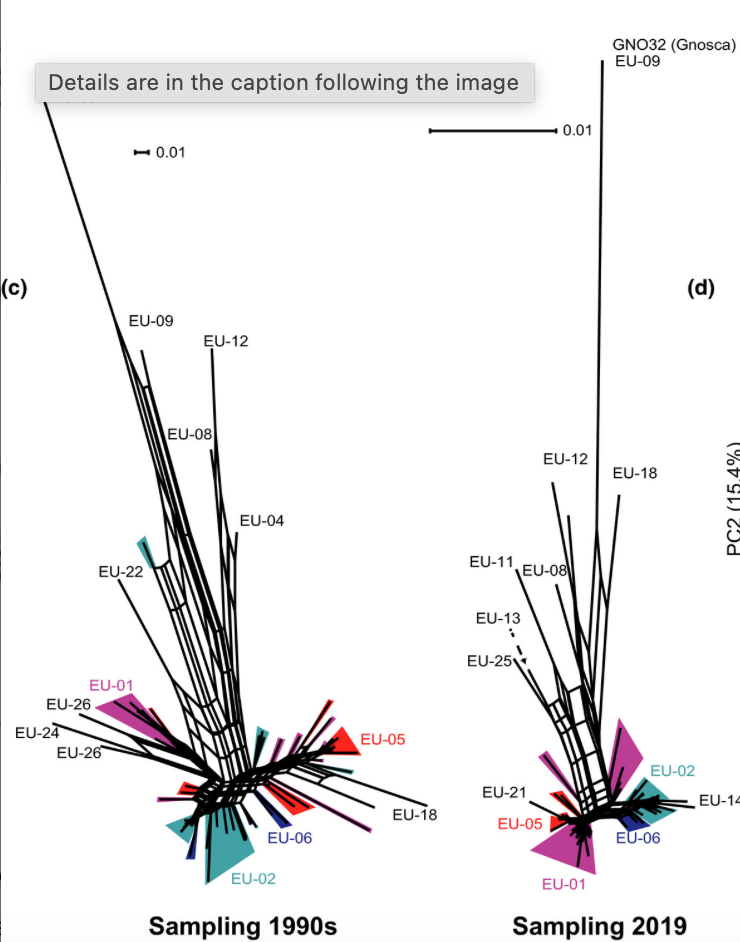
Temporal changes in pathogen diversity in a perennial plant–pathogen–hyperparasite system
Molecular Ecology
·
16 Feb 2022
·
doi:10.1111/mec.16386
Variability in an effector gene promoter of a necrotrophic fungal pathogen dictates epistasis and effector-triggered susceptibility in wheat
PLOS Pathogens
·
06 Jan 2022
·
doi:10.1371/journal.ppat.1010149

High-quality genome assembly of Pseudocercospora ulei the main threat to natural rubber trees
Genetics and Molecular Biology
·
01 Jan 2022
·
doi:10.1590/1678-4685-GMB-2021-0051
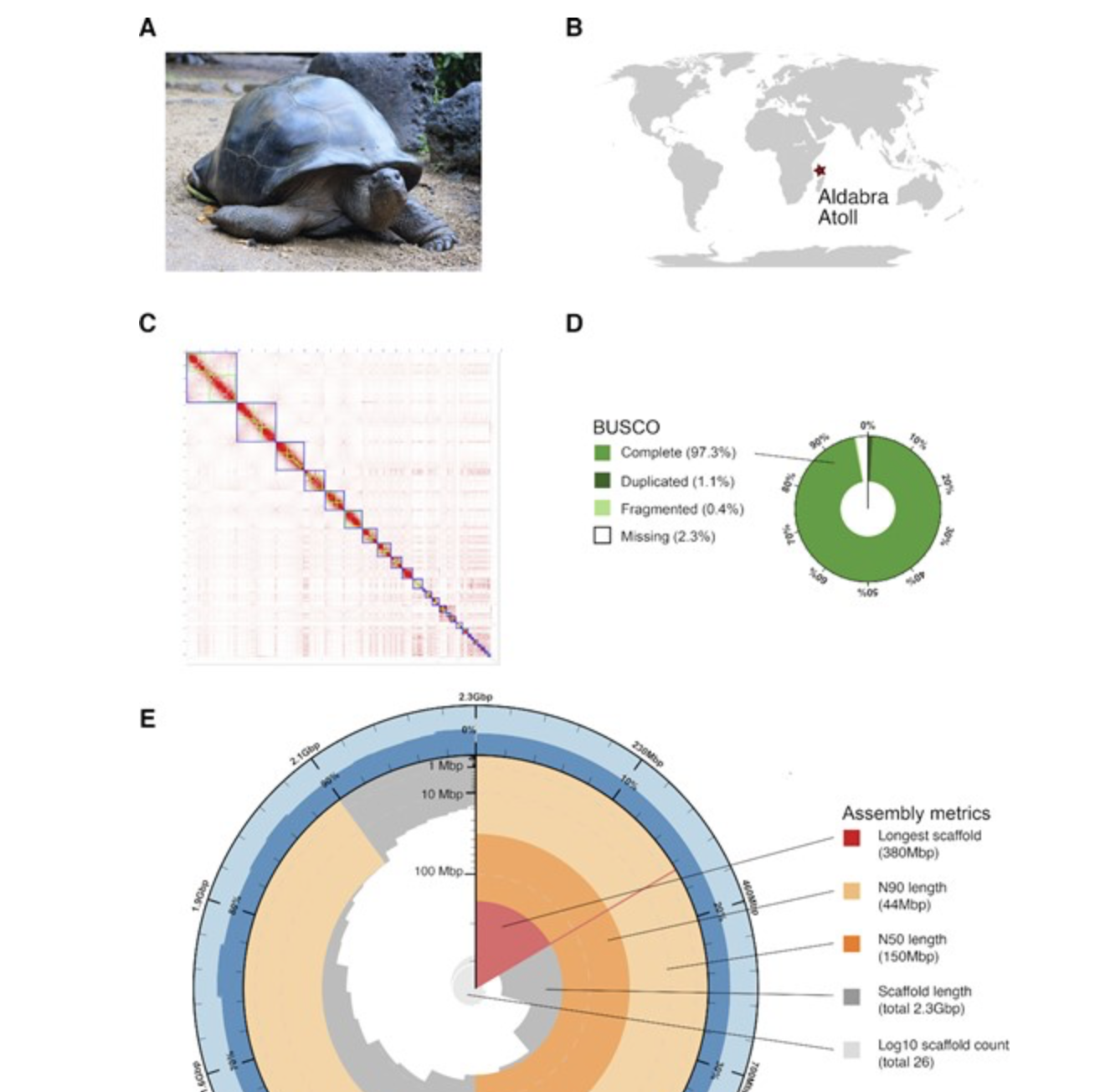
Chromosome-level genome assembly for the Aldabra giant tortoise enables insights into the genetic health of a threatened population
GigaScience
·
01 Jan 2022
·
doi:10.1093/gigascience/giac090
2021

Screening of grapevine red blotch virus in two European ampelographic collections
Journal of Plant Pathology
·
12 Nov 2021
·
doi:10.1007/s42161-021-00952-9
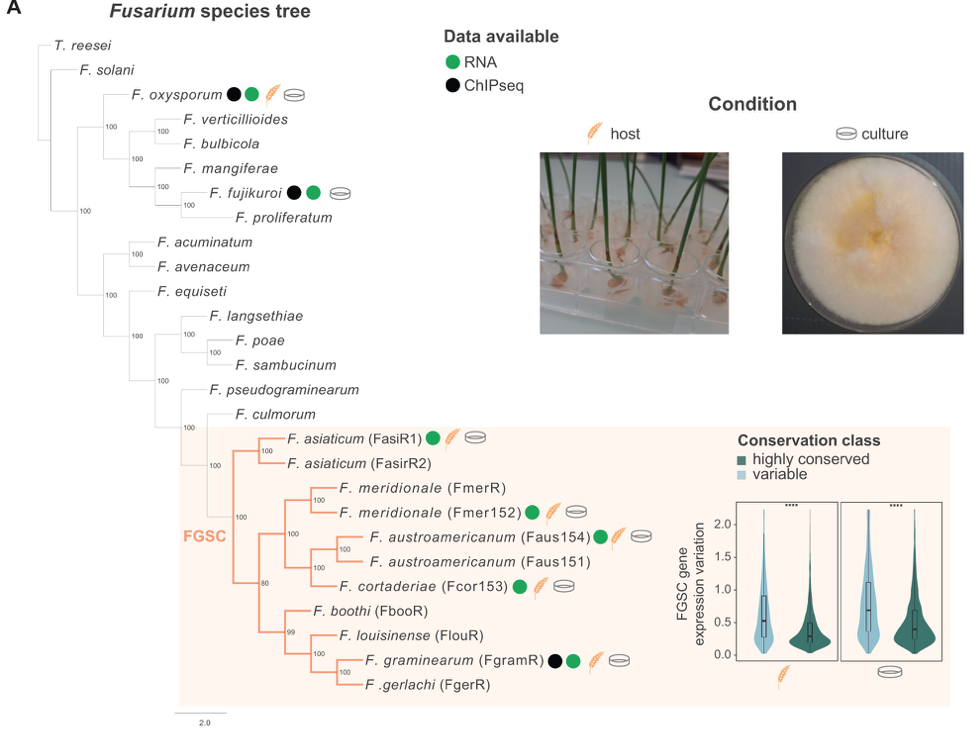
Histone H3K27 Methylation Perturbs Transcriptional Robustness and Underpins Dispensability of Highly Conserved Genes in Fungi
Molecular Biology and Evolution
·
09 Nov 2021
·
doi:10.1093/molbev/msab323
Population-level deep sequencing reveals the interplay of clonal and sexual reproduction in the fungal wheat pathogen Zymoseptoria tritici
Microbial Genomics
·
01 Nov 2021
·
doi:10.1099/mgen.0.000678

Soil composition and plant genotype determine benzoxazinoid‐mediated plant–soil feedbacks in cereals
Plant, Cell & Environment
·
23 Sep 2021
·
doi:10.1111/pce.14184
A population-level invasion by transposable elements triggers genome expansion in a fungal pathogen
eLife
·
16 Sep 2021
·
doi:10.7554/eLife.69249

Cryptic genetic structure and copy‐number variation in the ubiquitous forest symbiotic fungus Cenococcum geophilum
Environmental Microbiology
·
15 Sep 2021
·
doi:10.1111/1462-2920.15752
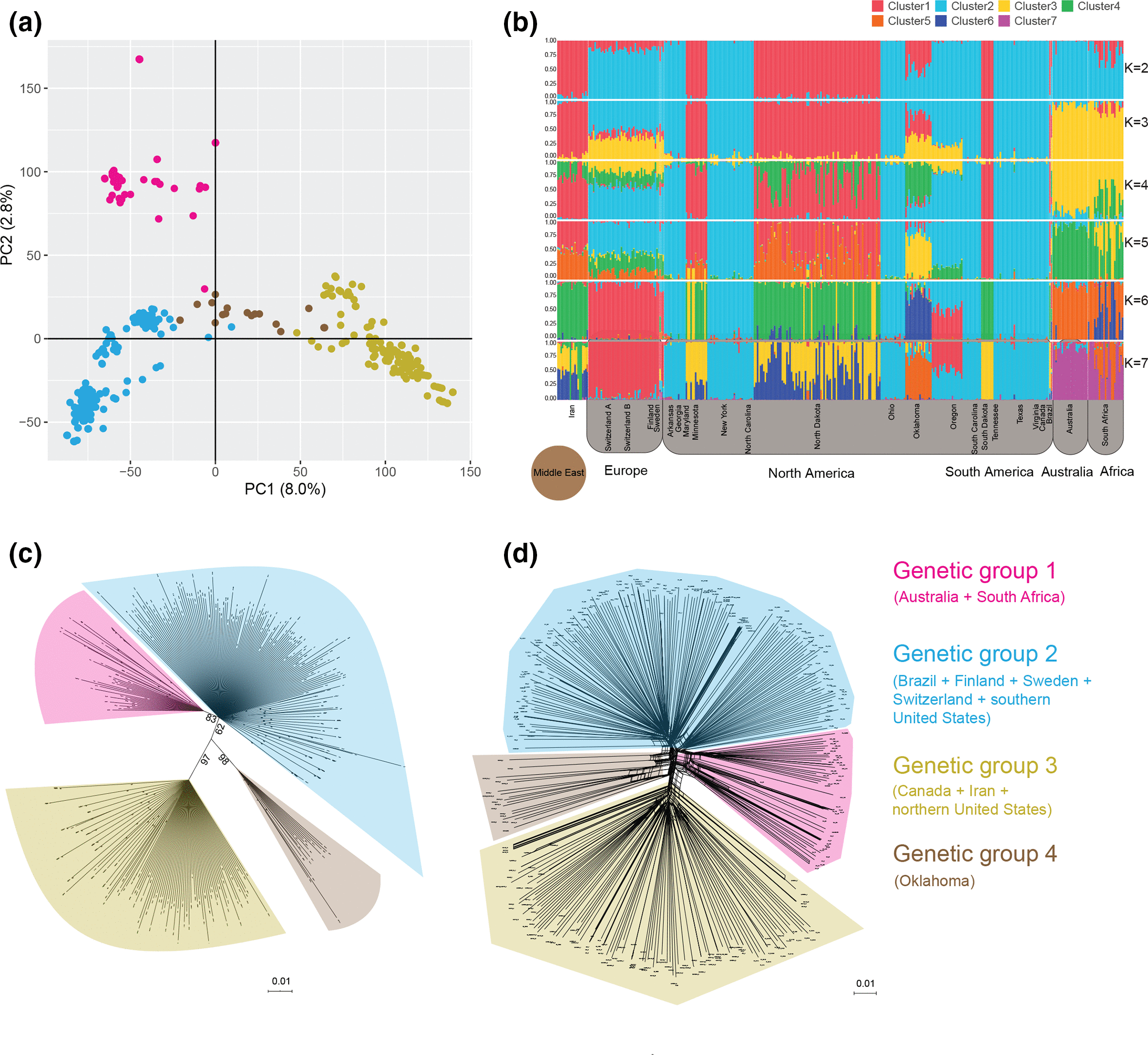
Population genomics of transposable element activation in the highly repressive genome of an agricultural pathogen
Microbial Genomics
·
31 Aug 2021
·
doi:10.1099/mgen.0.000540

Transcriptome‐wide SNPs for Botrychium lunaria ferns enable fine‐grained analysis of ploidy and population structure
Molecular Ecology Resources
·
07 Aug 2021
·
doi:10.1111/1755-0998.13478

A robust sequencing assay of a thousand amplicons for the high‐throughput population monitoring of Alpine ibex immunogenetics
Molecular Ecology Resources
·
07 Jul 2021
·
doi:10.1111/1755-0998.13452
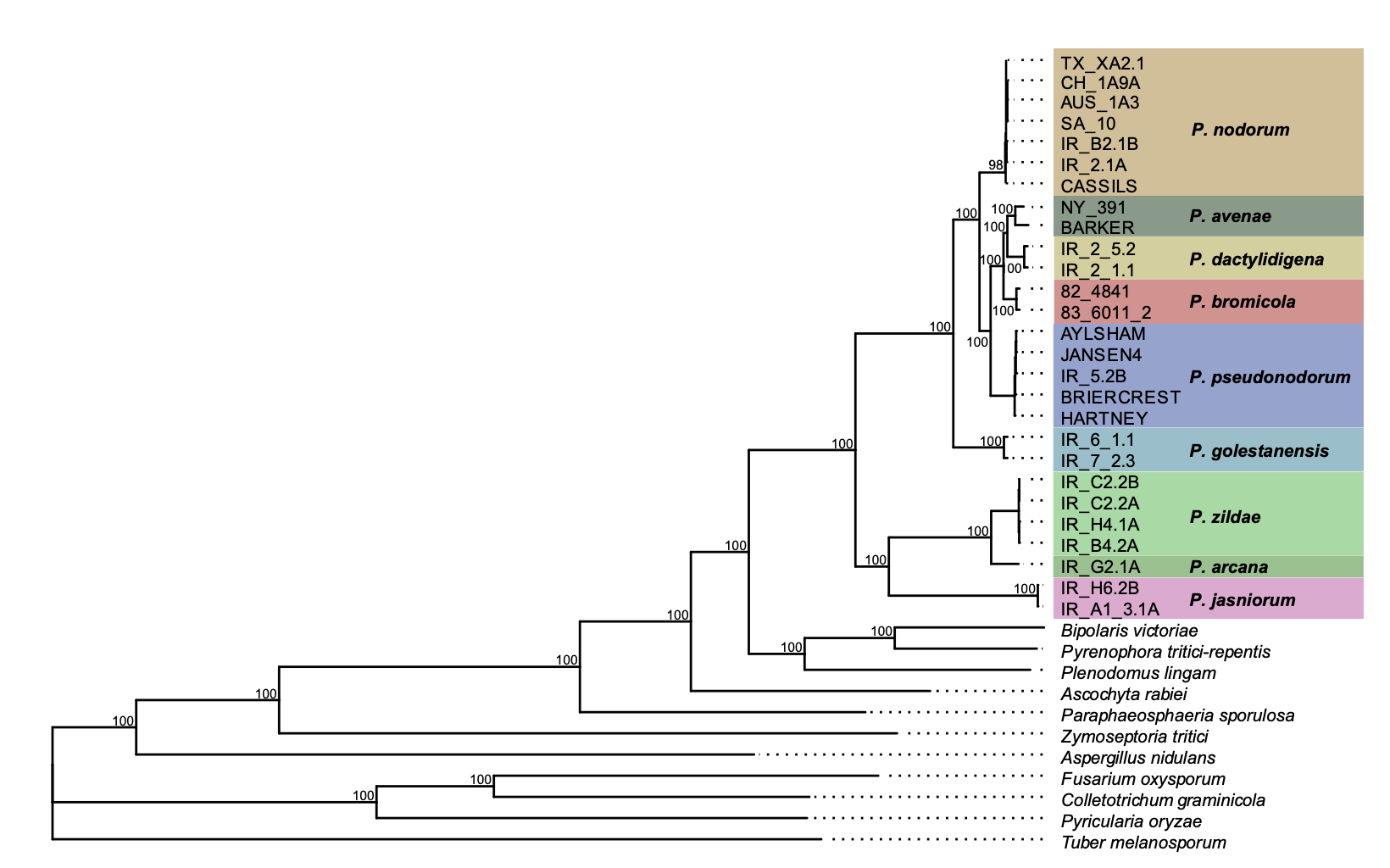
Genome-scale phylogenies reveal relationships among Parastagonospora species infecting domesticated and wild grasses
Persoonia - Molecular Phylogeny and Evolution of Fungi
·
30 Jun 2021
·
doi:10.3767/persoonia.2021.46.04
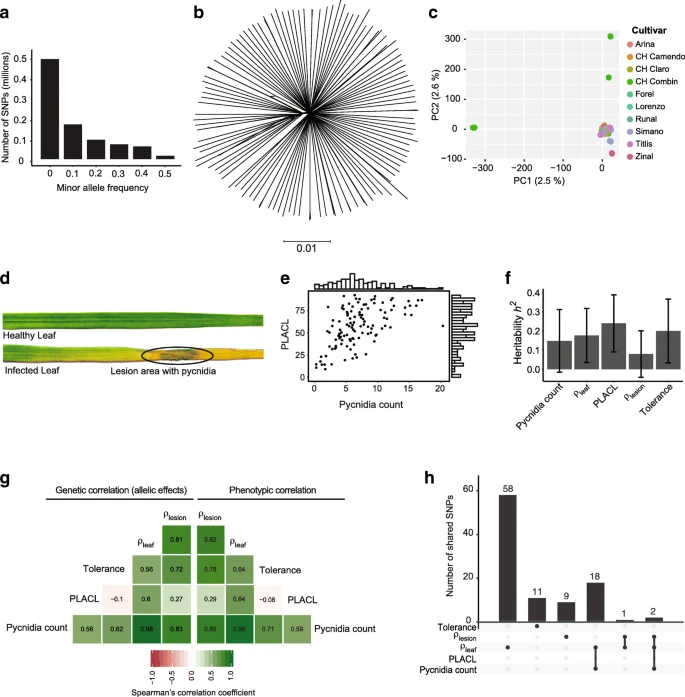
Machine-learning predicts genomic determinants of meiosis-driven structural variation in a eukaryotic pathogen
Nature Communications
·
10 Jun 2021
·
doi:10.1038/s41467-021-23862-x

A new method to determine the diet of pygmy hippopotamus in Taï National Park, Côte d’Ivoire
African Journal of Ecology
·
09 Jun 2021
·
doi:10.1111/aje.12888
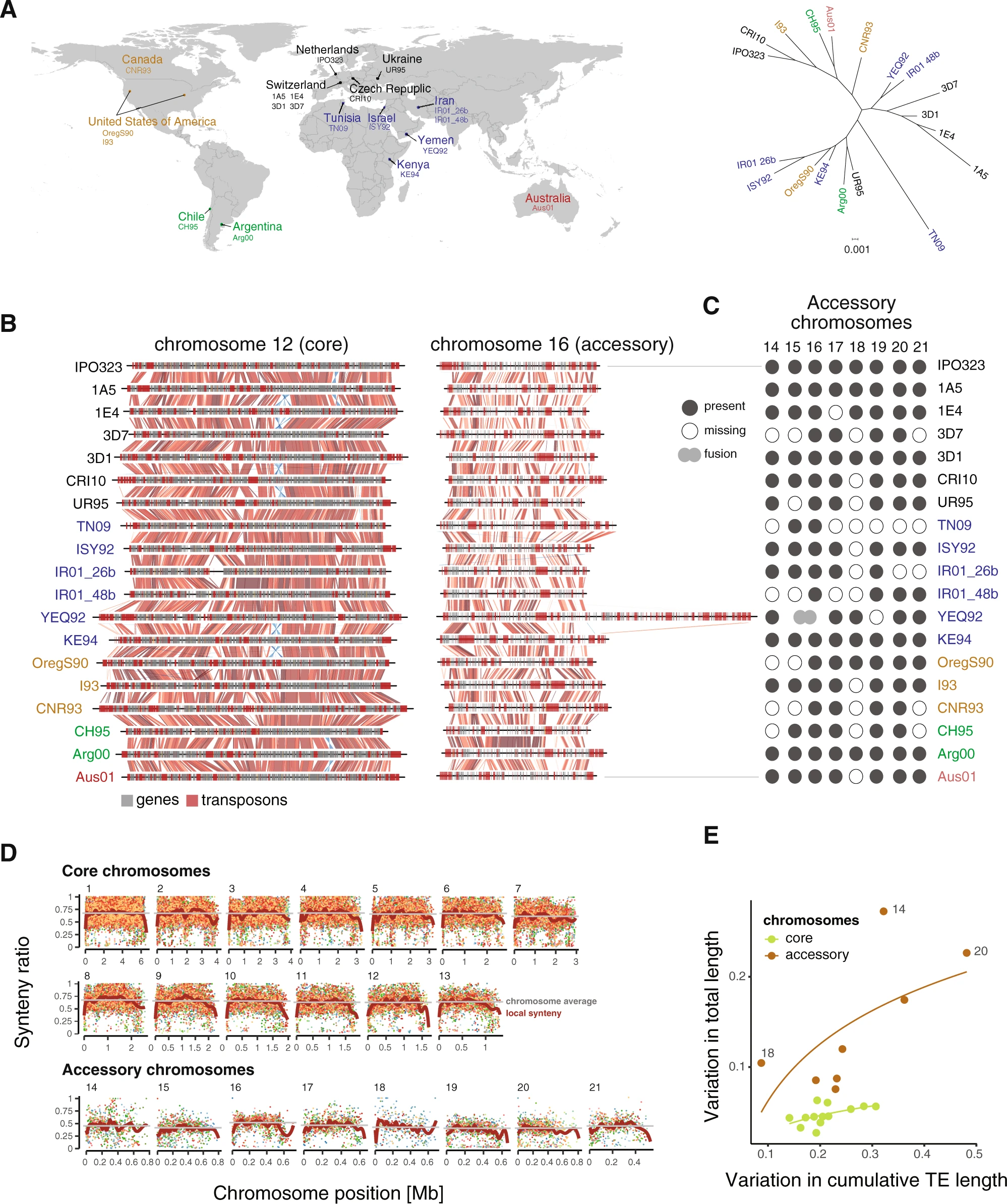
Rapid sequence evolution driven by transposable elements at a virulence locus in a fungal wheat pathogen
BMC Genomics
·
27 May 2021
·
doi:10.1186/s12864-021-07691-2
Genome-wide association study for septoria tritici blotch resistance reveals the occurrence and distribution of Stb6 in a historic Swiss landrace collection
Euphytica
·
11 May 2021
·
doi:10.1007/s10681-021-02843-7
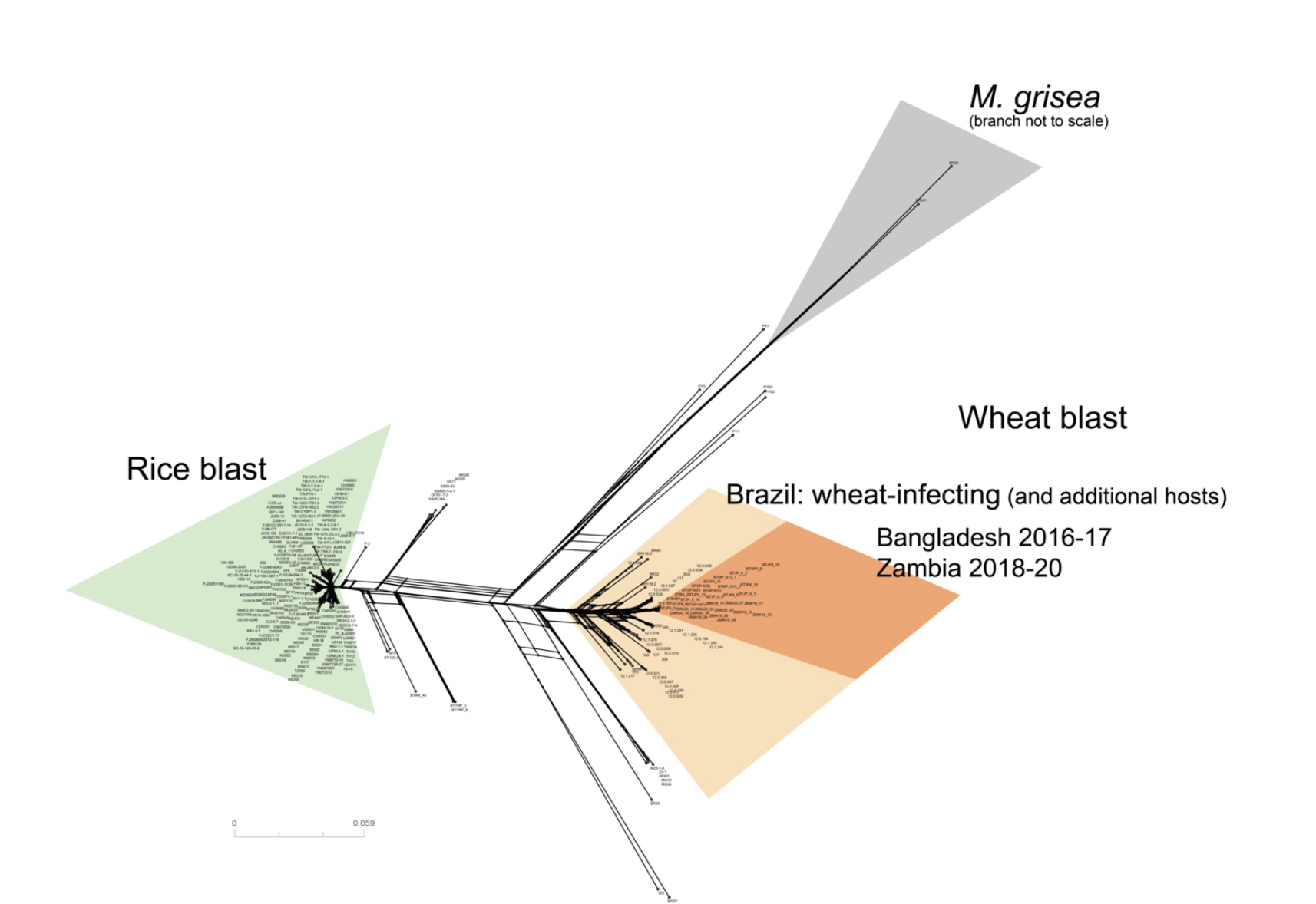
Whole-genome analyses of 286 Magnaporthe oryzae genomes suggest that an independent introduction of a global pandemic lineage is at the origin of the Zambia wheat blast outbreak
Zenodo
·
01 Apr 2021
·
doi:10.5281/zenodo.4655959
Emergence and diversification of a highly invasive chestnut pathogen lineage across southeastern Europe
eLife
·
05 Mar 2021
·
doi:10.7554/eLife.56279

Fusarium: more than a node or a foot-shaped basal cell
Studies in Mycology
·
01 Mar 2021
·
doi:10.1016/j.simyco.2021.100116
Mapping the adaptive landscape of a major agricultural pathogen reveals evolutionary constraints across heterogeneous environments
The ISME Journal
·
15 Jan 2021
·
doi:10.1038/s41396-020-00859-w
Chapter 10. Omics-based Detection, Identification and Quantification of GM Food and Feed: Current Challenges and Perspectives
Food Chemistry, Function and Analysis
·
01 Jan 2021
·
doi:10.1039/9781839163005-00257

Tackling microbial threats in agriculture with integrative imaging and computational approaches
Computational and Structural Biotechnology Journal
·
01 Jan 2021
·
doi:10.1016/j.csbj.2020.12.018

Transposable Elements in Fungi: Coevolution With the Host Genome Shapes, Genome Architecture, Plasticity and Adaptation
Encyclopedia of Mycology
·
01 Jan 2021
·
doi:10.1016/B978-0-12-819990-9.00042-1
2020

The complex genomic basis of rapid convergent adaptation to pesticides across continents in a fungal plant pathogen
Molecular Ecology
·
12 Dec 2020
·
doi:10.1111/mec.15737
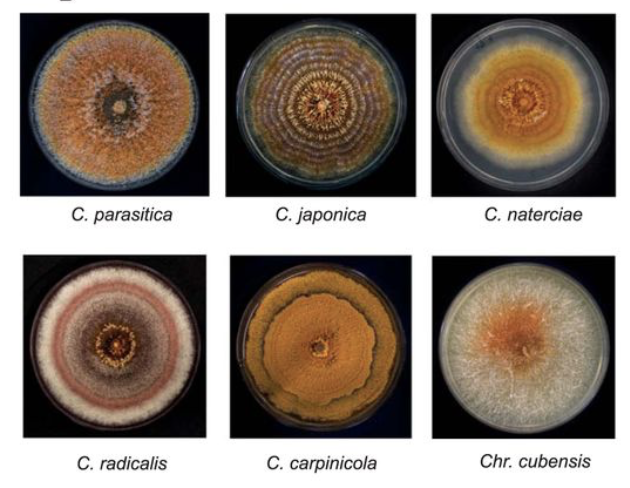
Comparative Genomics Analyses of Lifestyle Transitions at the Origin of an Invasive Fungal Pathogen in the GenusCryphonectria
mSphere
·
28 Oct 2020
·
doi:10.1128/mSphere.00737-20
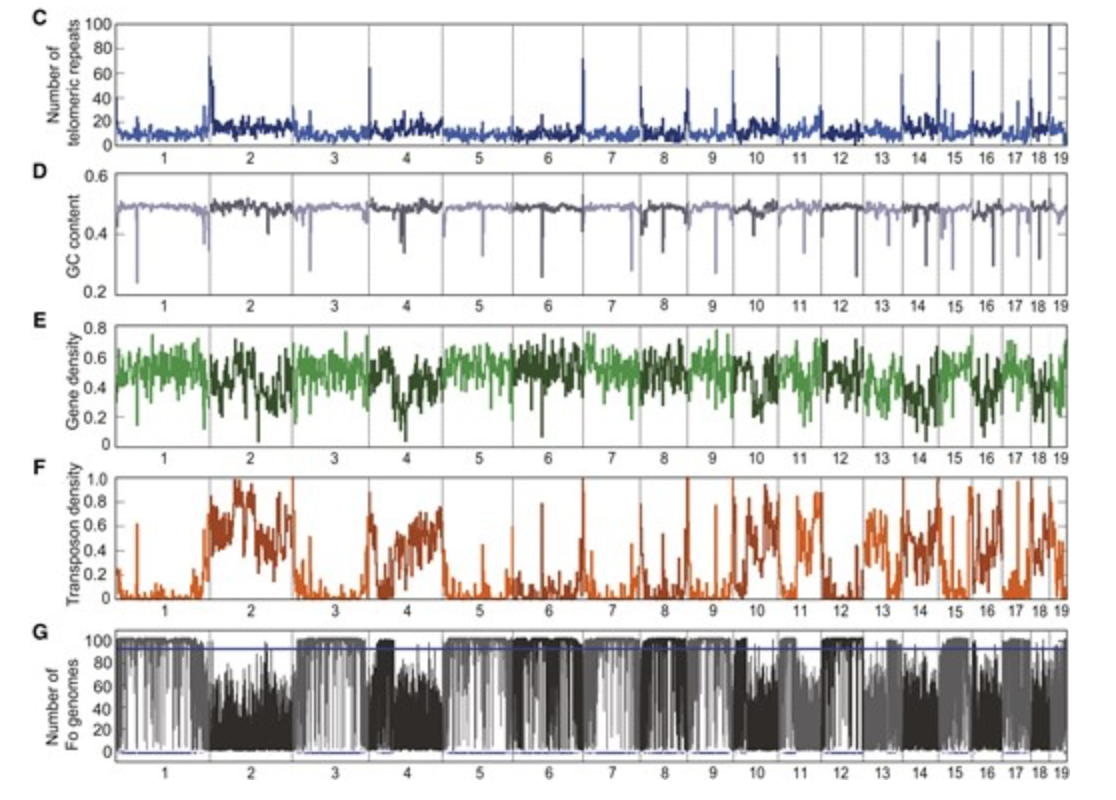
A Chromosome-Scale Genome Assembly for the Fusarium oxysporum Strain Fo5176 To Establish a Model Arabidopsis-Fungal Pathosystem
G3 Genes|Genomes|Genetics
·
01 Oct 2020
·
doi:10.1534/g3.120.401375
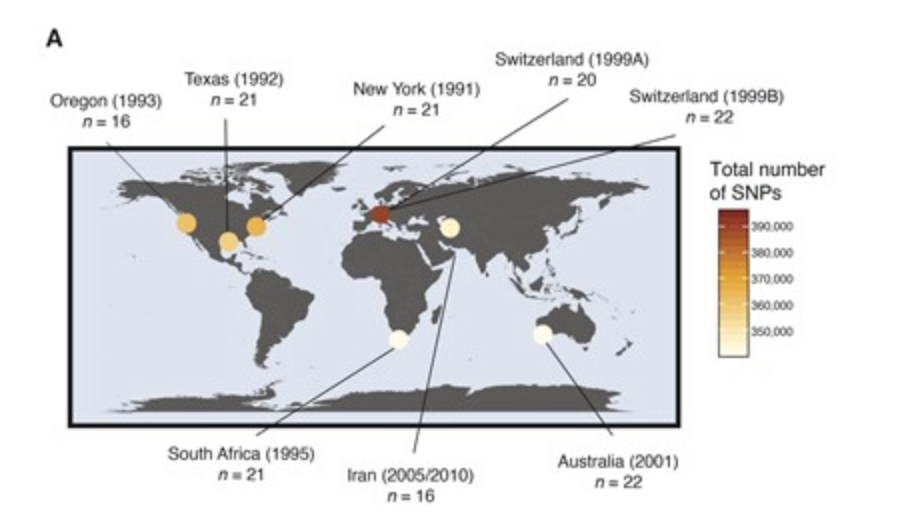
The Genetic Architecture of Emerging Fungicide Resistance in Populations of a Global Wheat Pathogen
Genome Biology and Evolution
·
28 Sep 2020
·
doi:10.1093/gbe/evaa203
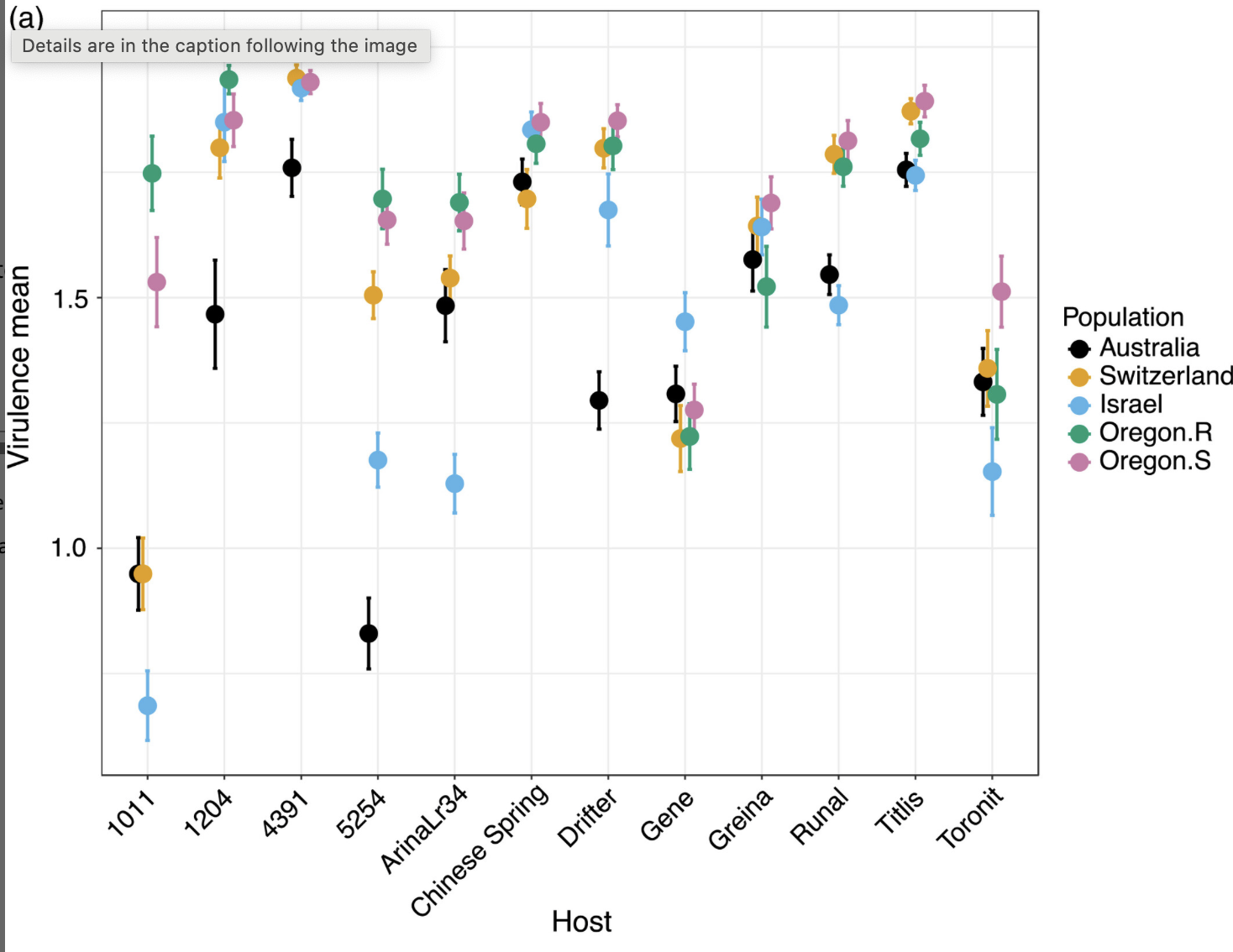
Maintenance of variation in virulence and reproduction in populations of an agricultural plant pathogen
Evolutionary Applications
·
24 Sep 2020
·
doi:10.1111/eva.13117
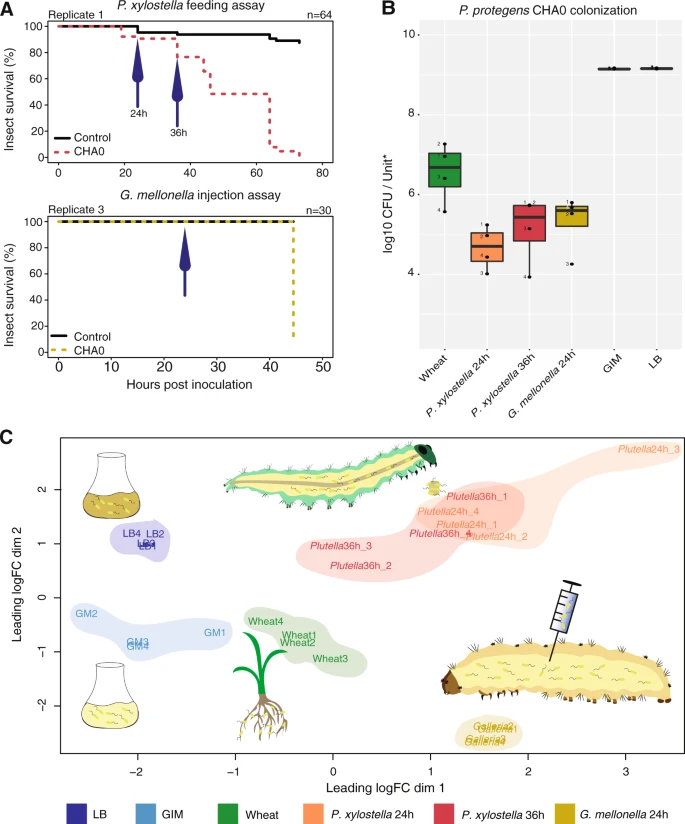
Transcriptome plasticity underlying plant root colonization and insect invasion by Pseudomonas protegens
The ISME Journal
·
02 Sep 2020
·
doi:10.1038/s41396-020-0729-9

Genome evolution in fungal plant pathogens: looking beyond the two-speed genome model
Fungal Biology Reviews
·
01 Sep 2020
·
doi:10.1016/j.fbr.2020.07.001
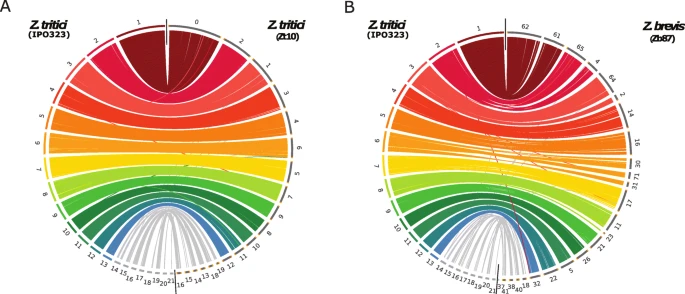
Genome compartmentalization predates species divergence in the plant pathogen genus Zymoseptoria
BMC Genomics
·
26 Aug 2020
·
doi:10.1186/s12864-020-06871-w

The rise and fall of genes: origins and functions of plant pathogen pangenomes
Current Opinion in Plant Biology
·
01 Aug 2020
·
doi:10.1016/j.pbi.2020.04.009

Natural selection drives population divergence for local adaptation in a wheat pathogen
Fungal Genetics and Biology
·
01 Aug 2020
·
doi:10.1016/j.fgb.2020.103398
A secreted LysM effector protects fungal hyphae through chitin-dependent homodimer polymerization
PLOS Pathogens
·
23 Jun 2020
·
doi:10.1371/journal.ppat.1008652
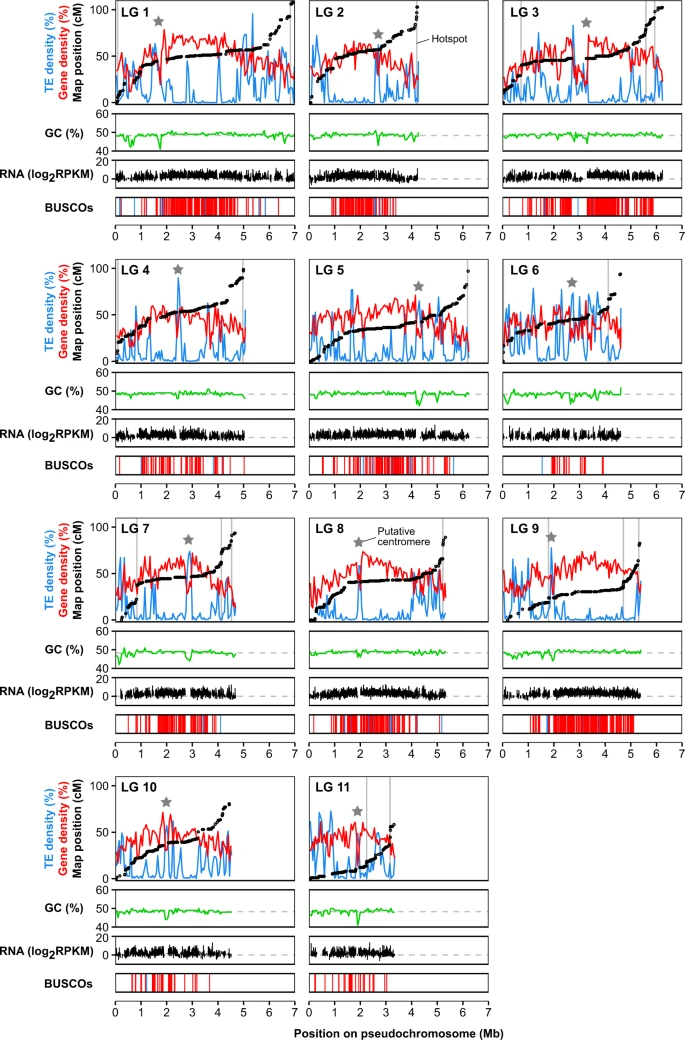
Chromosomal assembly and analyses of genome-wide recombination rates in the forest pathogenic fungus Armillaria ostoyae
Heredity
·
13 Mar 2020
·
doi:10.1038/s41437-020-0306-z
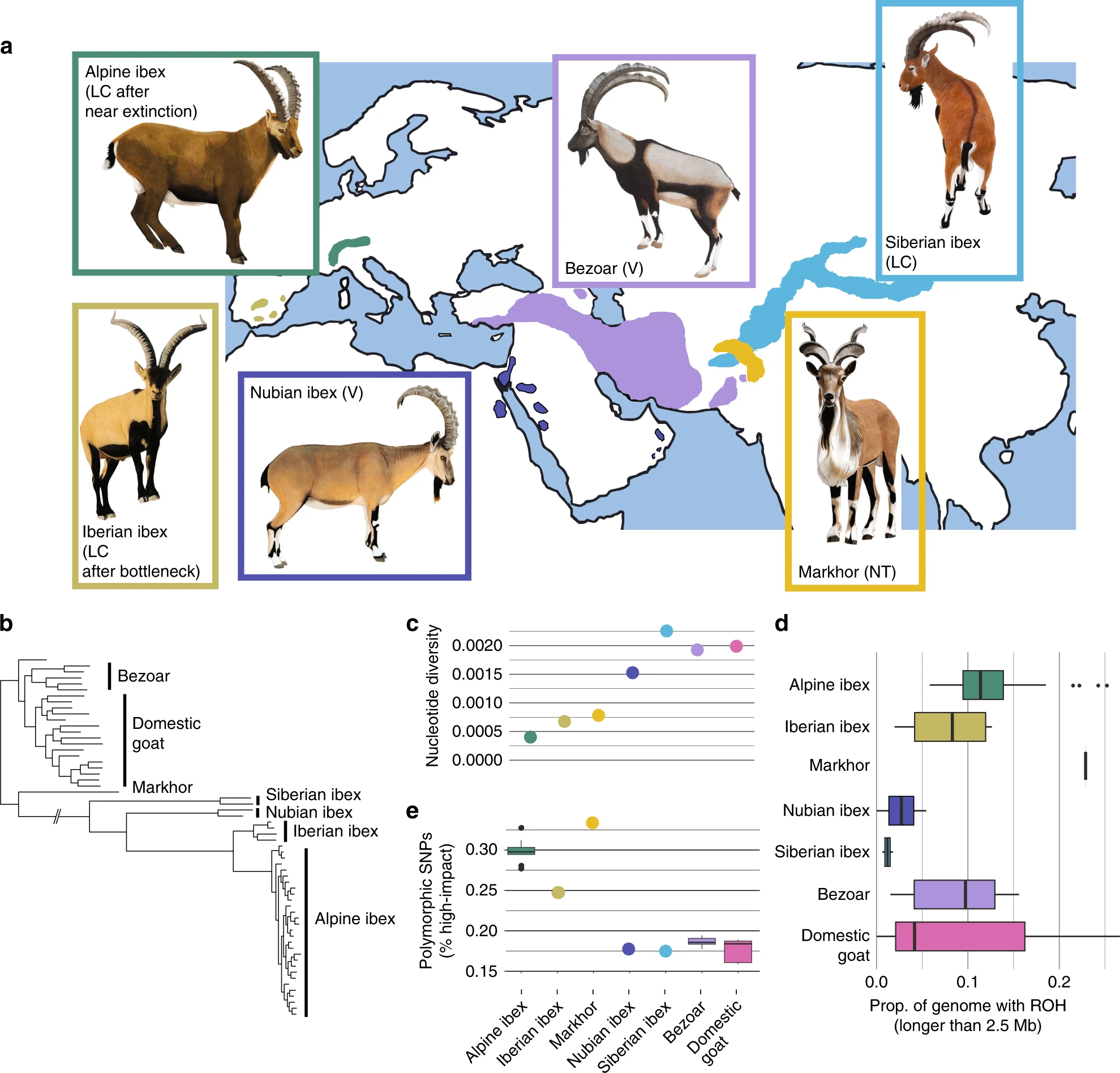
Purging of highly deleterious mutations through severe bottlenecks in Alpine ibex
Nature Communications
·
21 Feb 2020
·
doi:10.1038/s41467-020-14803-1
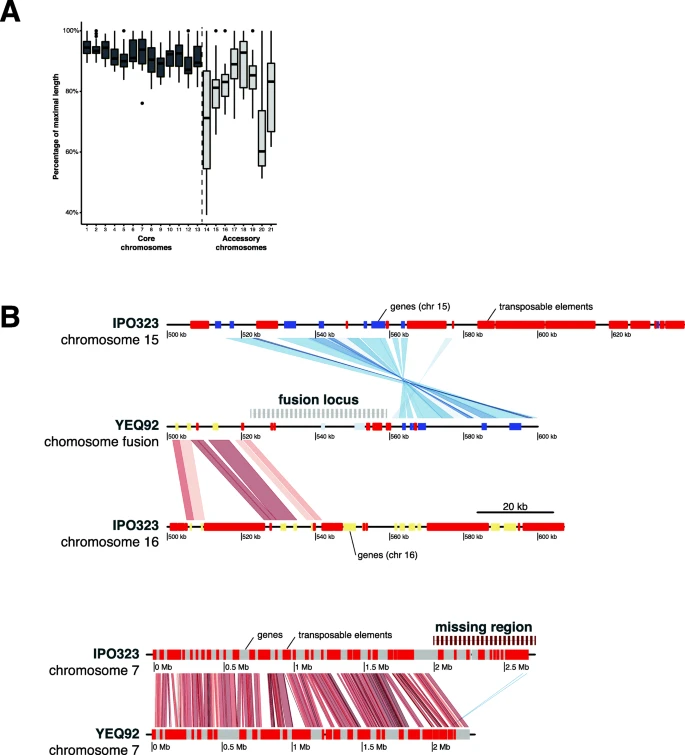
A 19-isolate reference-quality global pangenome for the fungal wheat pathogen Zymoseptoria tritici
BMC Biology
·
11 Feb 2020
·
doi:10.1186/s12915-020-0744-3
2019
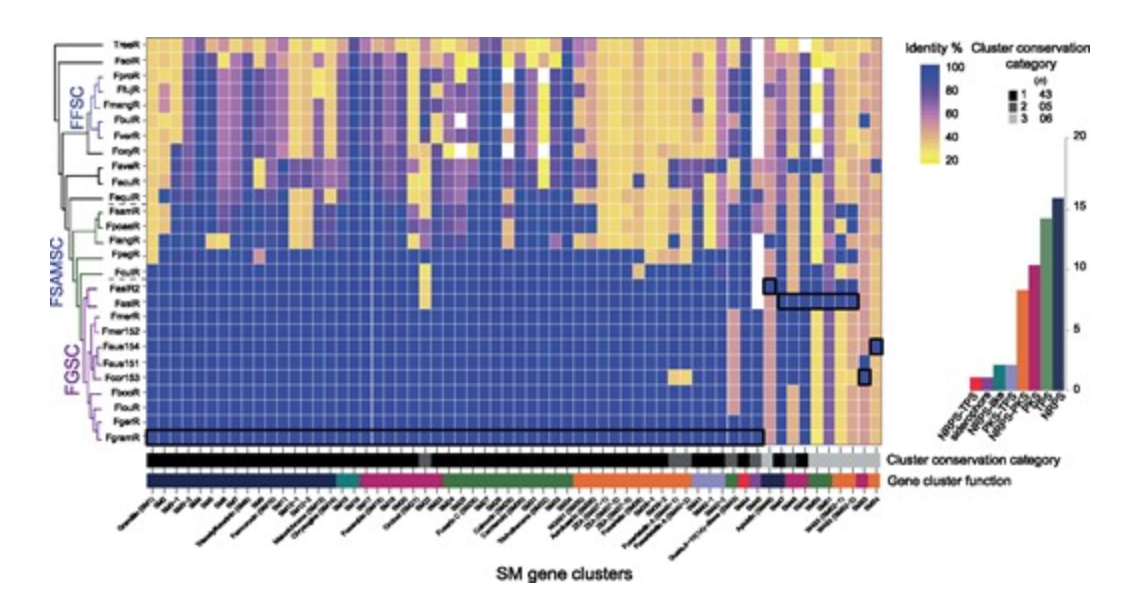
Complex Evolutionary Origins of Specialized Metabolite Gene Cluster Diversity among the Plant Pathogenic Fungi of the Fusarium graminearum Species Complex
Genome Biology and Evolution
·
14 Oct 2019
·
doi:10.1093/gbe/evz225
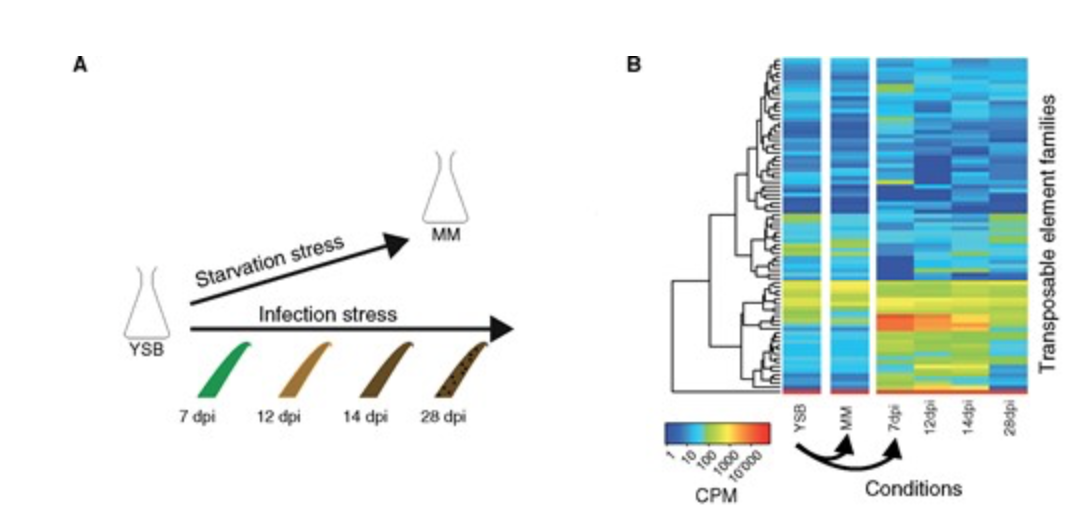
Stress-Driven Transposable Element De-repression Dynamics and Virulence Evolution in a Fungal Pathogen
Molecular Biology and Evolution
·
23 Sep 2019
·
doi:10.1093/molbev/msz216

The emergence of the multi‐species NIP1 effector in Rhynchosporium was accompanied by high rates of gene duplications and losses
Environmental Microbiology
·
11 Apr 2019
·
doi:10.1111/1462-2920.14583

Cautionary Notes on Use of the MoT3 Diagnostic Assay for Magnaporthe oryzae Wheat and Rice Blast Isolates
Phytopathology®
·
01 Apr 2019
·
doi:10.1094/PHYTO-06-18-0199-LE

Rapid Parallel Evolution of Azole Fungicide Resistance in Australian Populations of the Wheat Pathogen
Zymoseptoria tritici
Applied and Environmental Microbiology
·
15 Feb 2019
·
doi:10.1128/AEM.01908-18
2018

The Monothiol Glutaredoxin Grx4 Regulates Iron Homeostasis and Virulence in Cryptococcus neoformans
mBio
·
21 Dec 2018
·
doi:10.1128/mBio.02377-18

The birth and death of effectors in rapidly evolving filamentous pathogen genomes
Current Opinion in Microbiology
·
01 Dec 2018
·
doi:10.1016/j.mib.2018.01.020

Wheat blast: from its origins in South America to its emergence as a global threat
Molecular Plant Pathology
·
24 Oct 2018
·
doi:10.1111/mpp.12747
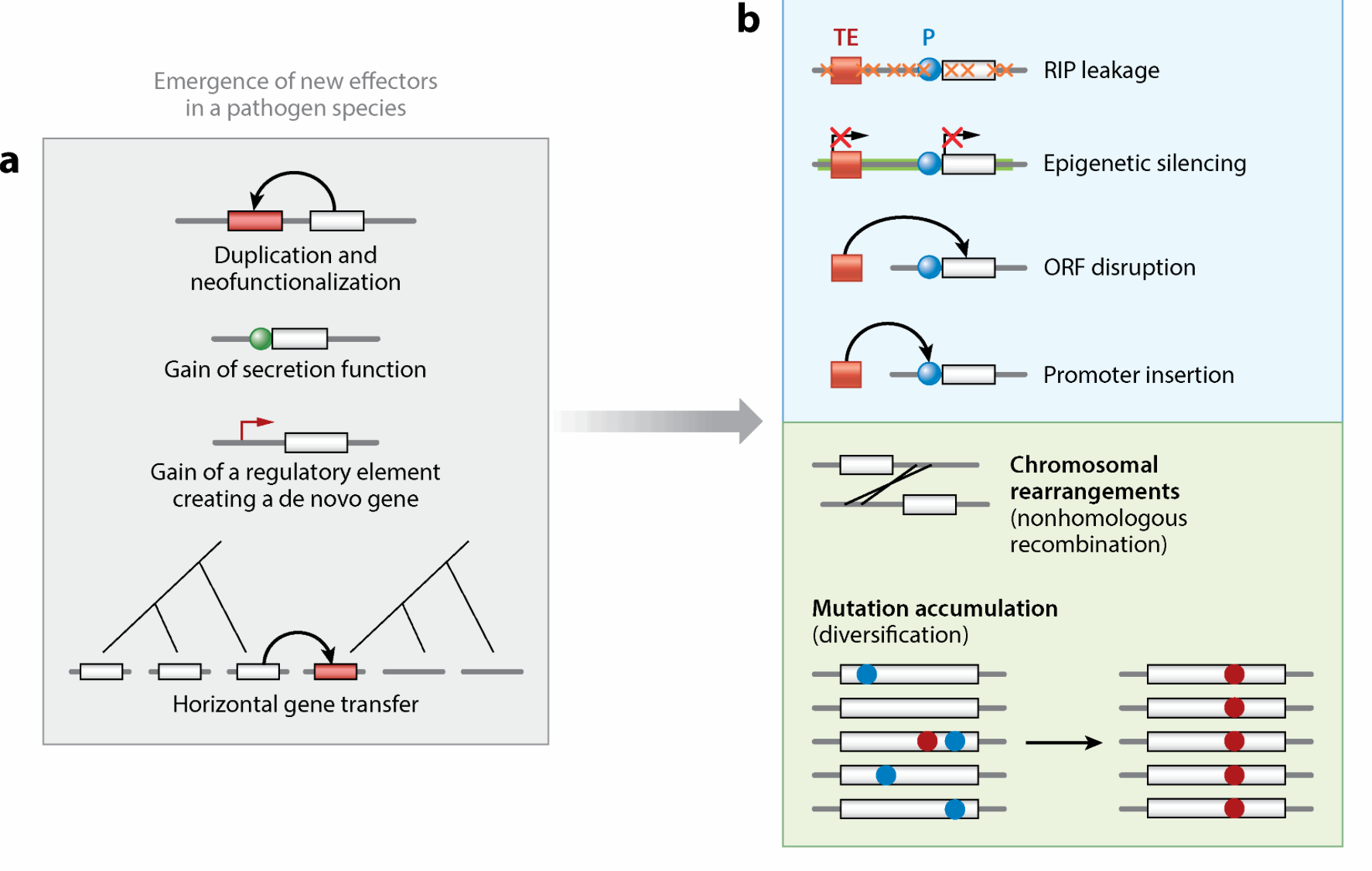
The Genome Biology of Effector Gene Evolution in Filamentous Plant Pathogens
Annual Review of Phytopathology
·
25 Aug 2018
·
doi:10.1146/annurev-phyto-080516-035303
Wheat Blast: Past, Present, and Future
Annual Review of Phytopathology
·
25 Aug 2018
·
doi:10.1146/annurev-phyto-080417-050036
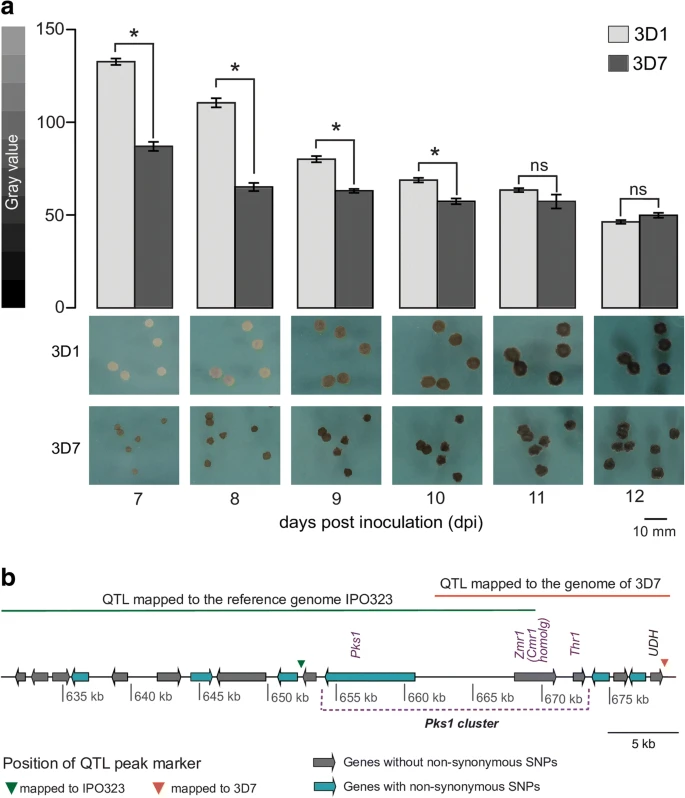
Transposable element insertions shape gene regulation and melanin production in a fungal pathogen of wheat
BMC Biology
·
16 Jul 2018
·
doi:10.1186/s12915-018-0543-2
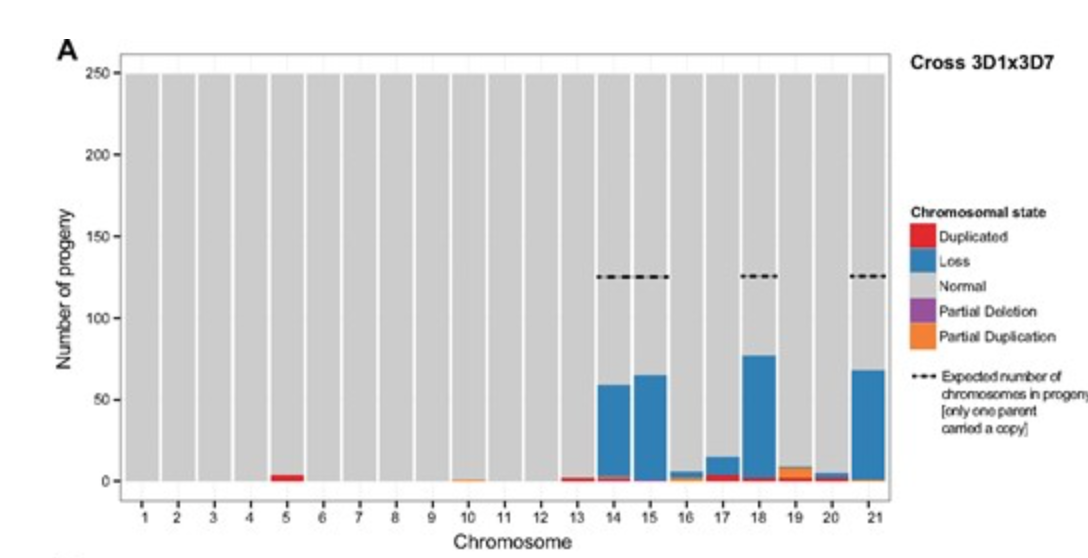
Meiosis Leads to Pervasive Copy-Number Variation and Distorted Inheritance of Accessory Chromosomes of the Wheat Pathogen Zymoseptoria tritici
Genome Biology and Evolution
·
29 May 2018
·
doi:10.1093/gbe/evy100

Genome‐wide evidence for divergent selection between populations of a major agricultural pathogen
Molecular Ecology
·
23 May 2018
·
doi:10.1111/mec.14711

Genome-Wide Detection of Genes Under Positive Selection in Worldwide Populations of the Barley Scald Pathogen
Genome Biology and Evolution
·
01 May 2018
·
doi:10.1093/gbe/evy087

Genomewide signatures of selection in Epichloë reveal candidate genes for host specialization
Molecular Ecology
·
27 Apr 2018
·
doi:10.1111/mec.14585

A fungal avirulence factor encoded in a highly plastic genomic region triggers partial resistance to septoria tritici blotch
New Phytologist
·
25 Apr 2018
·
doi:10.1111/nph.15180
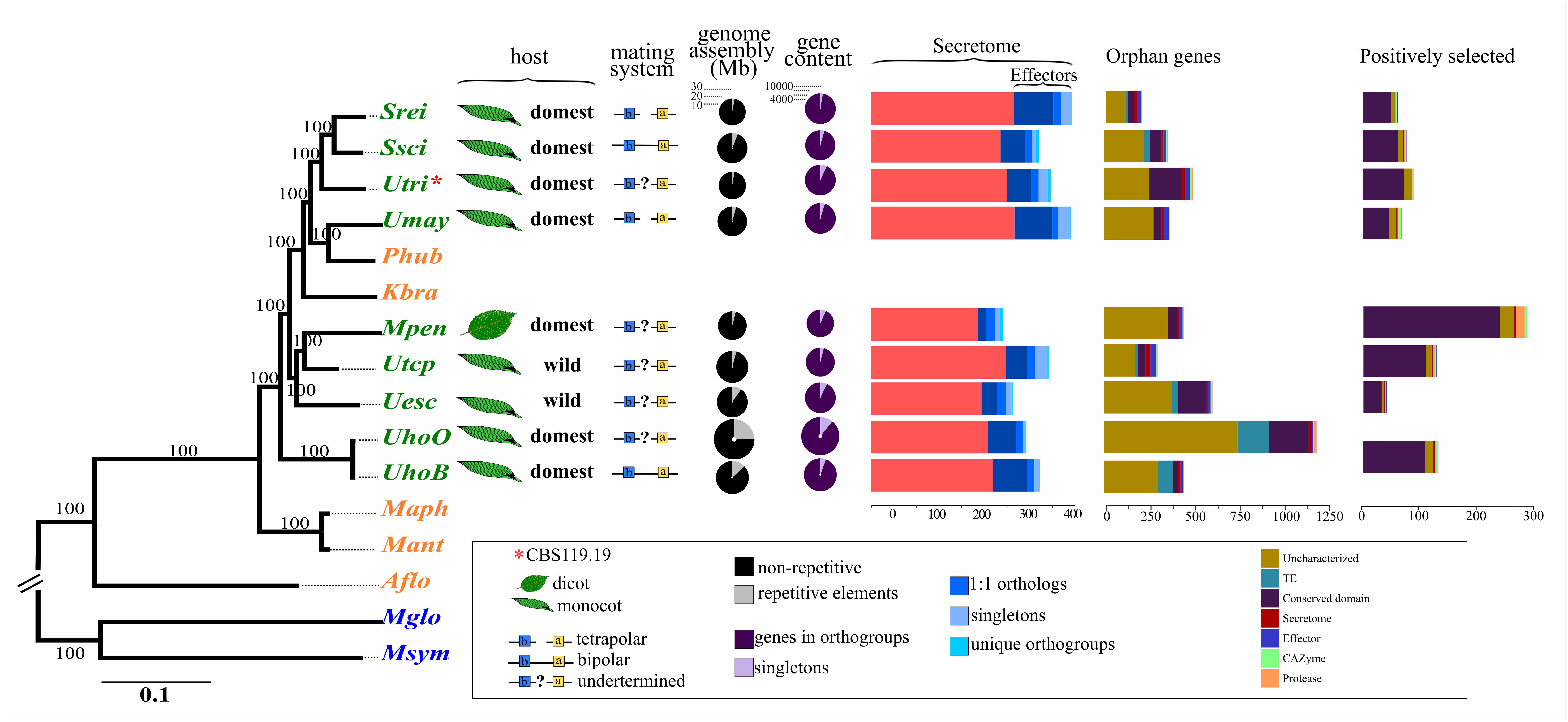
Comparative Genomics of Smut Pathogens: Insights From Orphans and Positively Selected Genes Into Host Specialization
Frontiers in Microbiology
·
06 Apr 2018
·
doi:10.3389/fmicb.2018.00660
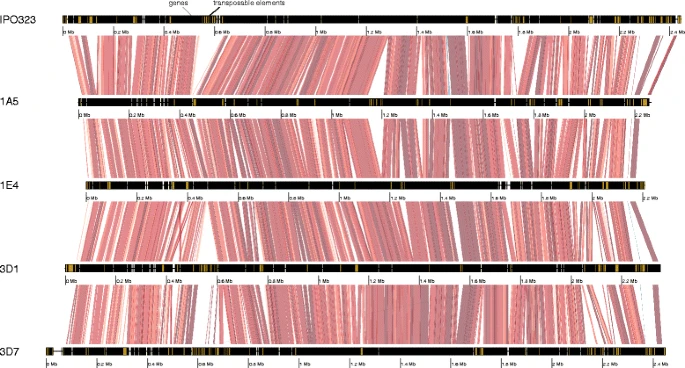
Pangenome analyses of the wheat pathogen Zymoseptoria tritici reveal the structural basis of a highly plastic eukaryotic genome
BMC Biology
·
11 Jan 2018
·
doi:10.1186/s12915-017-0457-4

Acetate provokes mitochondrial stress and cell death inUstilago maydis
Molecular Microbiology
·
03 Jan 2018
·
doi:10.1111/mmi.13894
2017

Nature's genetic screens: using genome‐wide association studies for effector discovery
Molecular Plant Pathology
·
11 Dec 2017
·
doi:10.1111/mpp.12592

High-density genetic mapping identifies the genetic basis of a natural colony morphology mutant in the root rot pathogen Armillaria ostoyae
Fungal Genetics and Biology
·
01 Nov 2017
·
doi:10.1016/j.fgb.2017.08.007

Population genomics analyses of European ibex species show lower diversity and higher inbreeding in reintroduced populations
Evolutionary Applications
·
27 Oct 2017
·
doi:10.1111/eva.12490

The wheat blast pathogenPyricularia graminis-triticihas complex origins and a disease cycle spanning multiple grass hosts
Cold Spring Harbor Laboratory
·
16 Oct 2017
·
doi:10.1101/203455

Distinct Trajectories of Massive Recent Gene Gains and Losses in Populations of a Microbial Eukaryotic Pathogen
Molecular Biology and Evolution
·
21 Jul 2017
·
doi:10.1093/molbev/msx208

Comparative Transcriptome Analyses inZymoseptoria triticiReveal Significant Differences in Gene Expression Among Strains During Plant Infection
Molecular Plant-Microbe Interactions®
·
01 Mar 2017
·
doi:10.1094/MPMI-07-16-0146-R

Can Evolution Supply What Ecology Demands?
Trends in Ecology & Evolution
·
01 Mar 2017
·
doi:10.1016/j.tree.2016.12.005

A small secreted protein in Zymoseptoria tritici is responsible for avirulence on wheat cultivars carrying the Stb6 resistance gene
New Phytologist
·
06 Feb 2017
·
doi:10.1111/nph.14434

Quantitative trait locus mapping reveals complex genetic architecture of quantitative virulence in the wheat pathogenZymoseptoria tritici
Molecular Plant Pathology
·
05 Feb 2017
·
doi:10.1111/mpp.12515
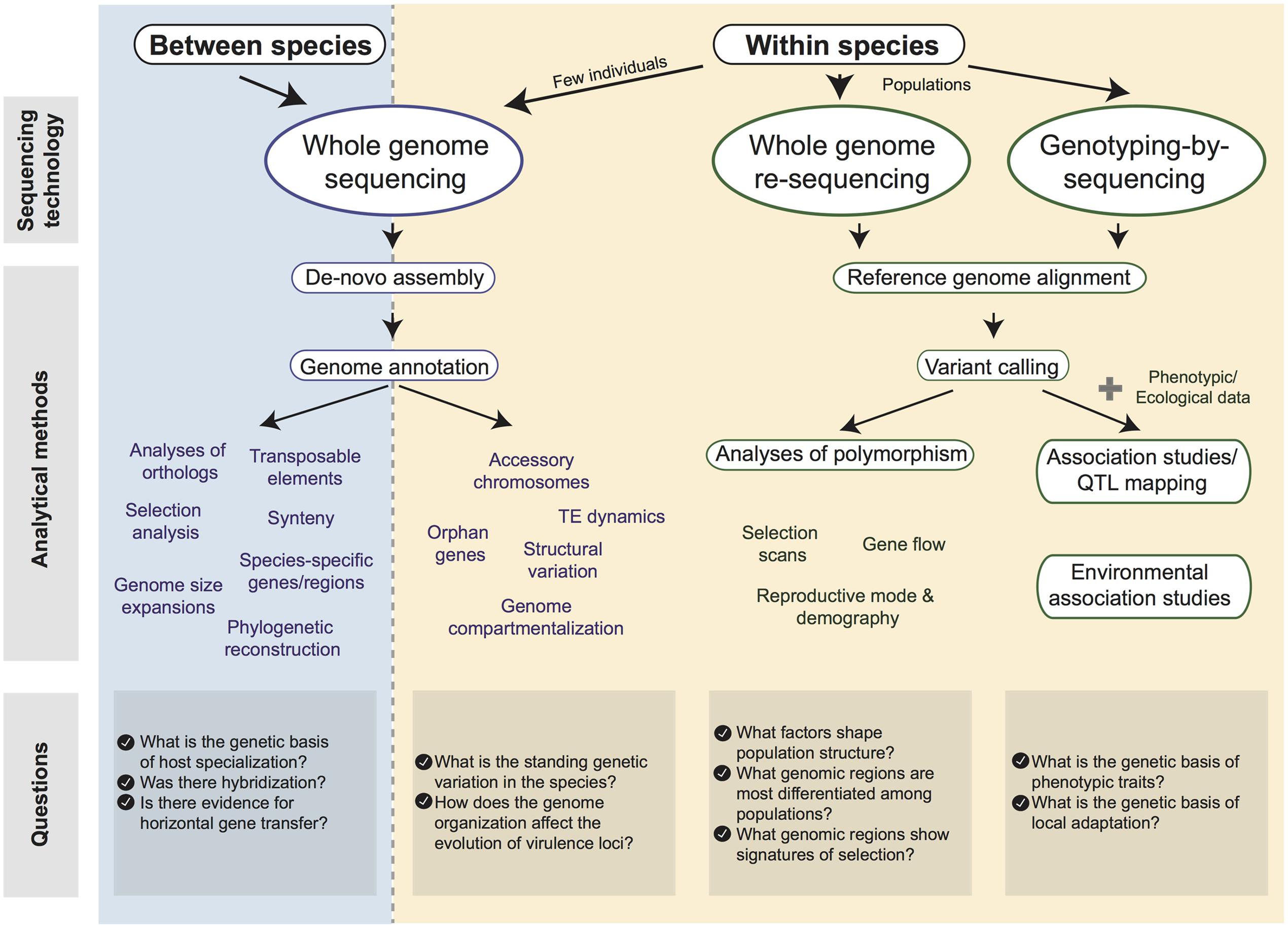
Using Population and Comparative Genomics to Understand the Genetic Basis of Effector-Driven Fungal Pathogen Evolution
Frontiers in Plant Science
·
03 Feb 2017
·
doi:10.3389/fpls.2017.00119
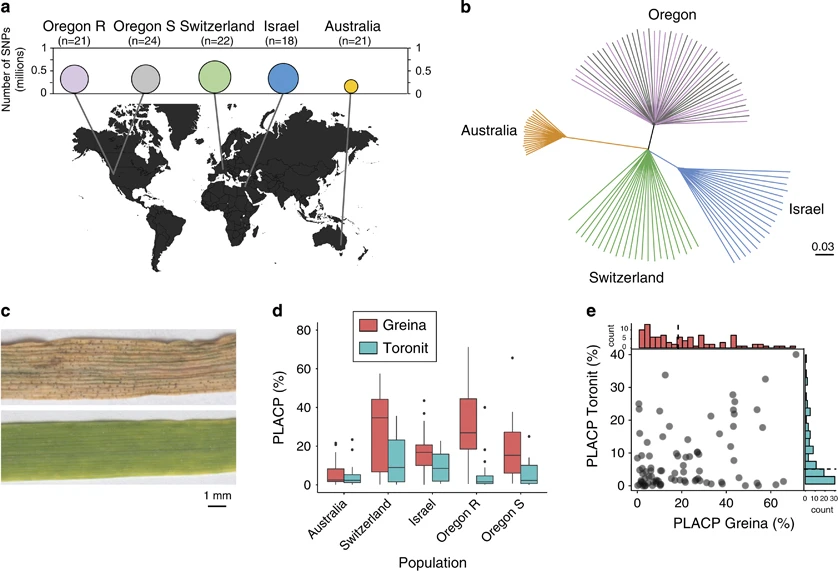
A fungal wheat pathogen evolved host specialization by extensive chromosomal rearrangements
The ISME Journal
·
24 Jan 2017
·
doi:10.1038/ismej.2016.196
2016

Multilocus resistance evolution to azole fungicides in fungal plant pathogen populations
Molecular Ecology
·
30 Nov 2016
·
doi:10.1111/mec.13916

Maize susceptibility to Ustilago maydis is influenced by genetic and chemical perturbation of carbohydrate allocation
Molecular Plant Pathology
·
14 Nov 2016
·
doi:10.1111/mpp.12486

Chloroplast‐associated metabolic functions influence the susceptibility of maize to Ustilago maydis
Molecular Plant Pathology
·
14 Nov 2016
·
doi:10.1111/mpp.12485

The Evolution of Orphan Regions in Genomes of a Fungal Pathogen of Wheat
mBio
·
02 Nov 2016
·
doi:10.1128/mBio.01231-16

The genetic basis of local adaptation for pathogenic fungi in agricultural ecosystems
Molecular Ecology
·
24 Oct 2016
·
doi:10.1111/mec.13870

What the population genetic structures of host and pathogen tell us about disease evolution
New Phytologist
·
13 Oct 2016
·
doi:10.1111/nph.14203
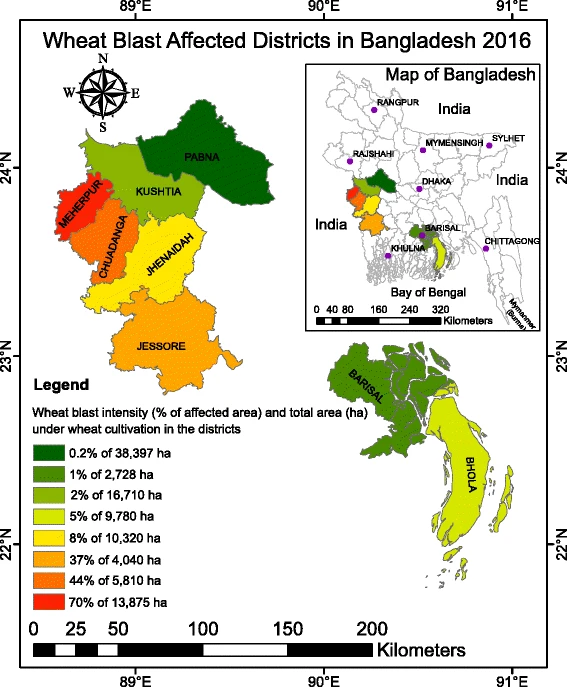
Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae
BMC Biology
·
03 Oct 2016
·
doi:10.1186/s12915-016-0309-7

Evolution in the absence of sex: Ideas revisited in the post‐genomics age (retrospective on DOI 10.1002/bies.201300155)
BioEssays
·
19 Sep 2016
·
doi:10.1002/bies.201600195

Strong reproductive isolation and narrow genomic tracts of differentiation among three woodpecker species in secondary contact
Molecular Ecology
·
26 Aug 2016
·
doi:10.1111/mec.13751

The putative phospholipase Lip2 counteracts oxidative damage and influences the virulence of Ustilago maydis
Molecular Plant Pathology
·
21 Apr 2016
·
doi:10.1111/mpp.12391

Analysis of the Protein Kinase A-Regulated Proteome of Cryptococcus neoformans Identifies a Role for the Ubiquitin-Proteasome Pathway in Capsule Formation
mBio
·
02 Mar 2016
·
doi:10.1128/mBio.01862-15

Comparative transcriptomic analyses of Z ymoseptoria tritici strains show complex lifestyle transitions and intraspecific variability in transcription profiles
Molecular Plant Pathology
·
08 Feb 2016
·
doi:10.1111/mpp.12333
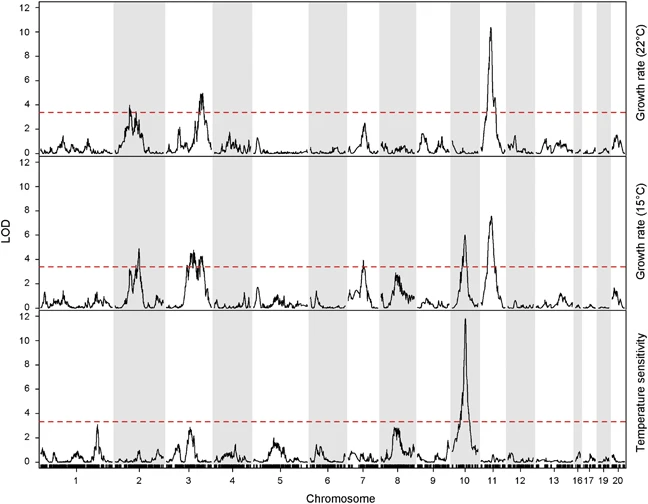
QTL mapping of temperature sensitivity reveals candidate genes for thermal adaptation and growth morphology in the plant pathogenic fungus Zymoseptoria tritici
Heredity
·
13 Jan 2016
·
doi:10.1038/hdy.2015.111
2015
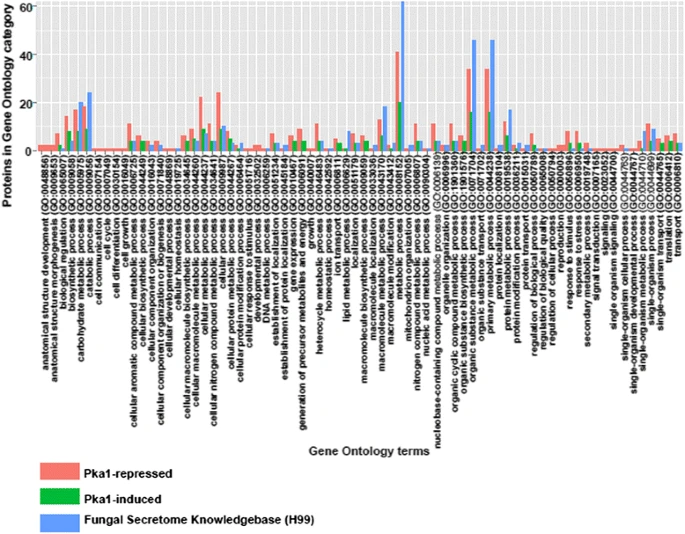
Secretome profiling of Cryptococcus neoformans reveals regulation of a subset of virulence-associated proteins and potential biomarkers by protein kinase A
BMC Microbiology
·
09 Oct 2015
·
doi:10.1186/s12866-015-0532-3

The Impact of Recombination Hotspots on Genome Evolution of a Fungal Plant Pathogen
Genetics
·
21 Sep 2015
·
doi:10.1534/genetics.115.180968

QTL mapping of fungicide sensitivity reveals novel genes and pleiotropy with melanization in the pathogen Zymoseptoria tritici
Fungal Genetics and Biology
·
01 Jul 2015
·
doi:10.1016/j.fgb.2015.05.001

Analysis of the nasal vestibule mycobiome in patients with allergic rhinitis
Mycoses
·
10 Feb 2015
·
doi:10.1111/myc.12296
2014

Quantitative Trait Locus Mapping of Melanization in the Plant Pathogenic FungusZymoseptoria tritici
G3 Genes|Genomes|Genetics
·
01 Dec 2014
·
doi:10.1534/g3.114.015289

Highly Recombinant VGII Cryptococcus gattii Population Develops Clonal Outbreak Clusters through both Sexual Macroevolution and Asexual Microevolution
mBio
·
29 Aug 2014
·
doi:10.1128/mBio.01494-14
Introgression from Domestic Goat Generated Variation at the Major Histocompatibility Complex of Alpine Ibex
PLoS Genetics
·
19 Jun 2014
·
doi:10.1371/journal.pgen.1004438

The evolving fungal genome
Fungal Biology Reviews
·
01 May 2014
·
doi:10.1016/j.fbr.2014.02.001

Hitchhiking Selection Is Driving Intron Gain in a Pathogenic Fungus
Molecular Biology and Evolution
·
07 Apr 2014
·
doi:10.1093/molbev/msu123

Learn from the fungi: Adaptive evolution without sex in fungal pathogens (comment on DOI 10.1002/bies.201300155)
BioEssays
·
05 Mar 2014
·
doi:10.1002/bies.201400030
2013

An assay for quantitative virulence in R hynchosporium commune reveals an association between effector genotype and virulence
Plant Pathology
·
06 Aug 2013
·
doi:10.1111/ppa.12111
Breakage-fusion-bridge Cycles and Large Insertions Contribute to the Rapid Evolution of Accessory Chromosomes in a Fungal Pathogen
PLoS Genetics
·
13 Jun 2013
·
doi:10.1371/journal.pgen.1003567

Coevolution and Life Cycle Specialization of Plant Cell Wall Degrading Enzymes in a Hemibiotrophic Pathogen
Molecular Biology and Evolution
·
20 Mar 2013
·
doi:10.1093/molbev/mst041
2012
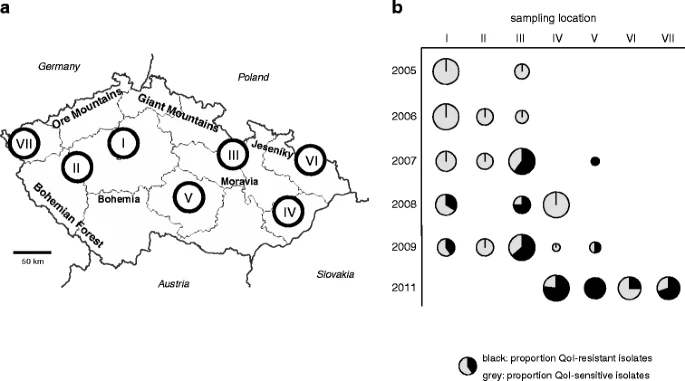
Population genetic structure of Mycosphaerella graminicola and Quinone Outside Inhibitor (QoI) resistance in the Czech Republic
European Journal of Plant Pathology
·
14 Sep 2012
·
doi:10.1007/s10658-012-0080-8
The Accessory Genome as a Cradle for Adaptive Evolution in Pathogens
PLoS Pathogens
·
26 Apr 2012
·
doi:10.1371/journal.ppat.1002608
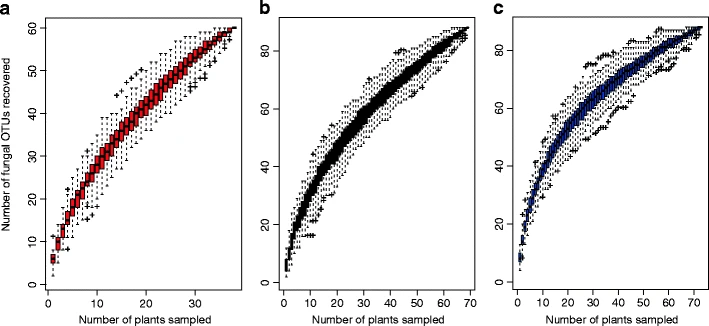
What if esca disease of grapevine were not a fungal disease?
Fungal Diversity
·
24 Apr 2012
·
doi:10.1007/s13225-012-0171-z

Intron Gains and Losses in the Evolution of Fusarium and Cryptococcus Fungi
Genome Biology and Evolution
·
01 Jan 2012
·
doi:10.1093/gbe/evs091
2011

Evidence for Extensive Recent Intron Transposition in Closely Related Fungi
Current Biology
·
01 Dec 2011
·
doi:10.1016/j.cub.2011.10.041

The transcriptome of the arbuscular mycorrhizal fungus Glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont
New Phytologist
·
16 Nov 2011
·
doi:10.1111/j.1469-8137.2011.03948.x

A cryptic heterogametic transition revealed by sex‐linked DNA markers in Palearctic green toads
Journal of Evolutionary Biology
·
21 Feb 2011
·
doi:10.1111/j.1420-9101.2011.02239.x
2010

Arbuscular Mycorrhiza: The Challenge to Understand the Genetics of the Fungal Partner
Annual Review of Genetics
·
01 Dec 2010
·
doi:10.1146/annurev-genet-102108-134239

Segregation in a Mycorrhizal Fungus Alters Rice Growth and Symbiosis-Specific Gene Transcription
Current Biology
·
01 Jul 2010
·
doi:10.1016/j.cub.2010.05.031
2009

High-Level Molecular Diversity of Copper-Zinc Superoxide Dismutase Genes among and within Species of Arbuscular Mycorrhizal Fungi
Applied and Environmental Microbiology
·
01 Apr 2009
·
doi:10.1128/AEM.01974-08
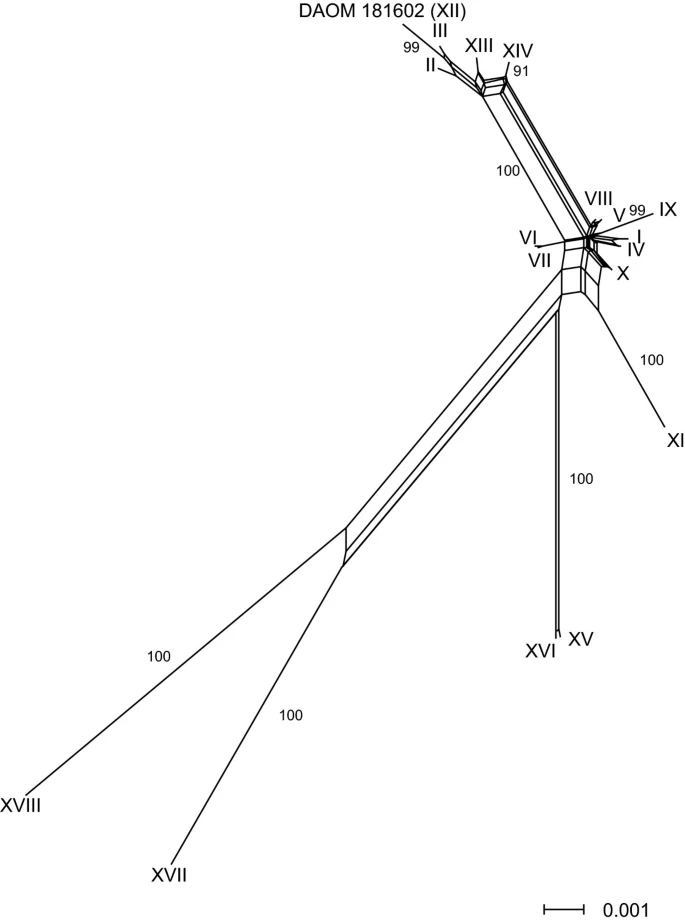
Recombination in Glomus intraradices, a supposed ancient asexual arbuscular mycorrhizal fungus
BMC Evolutionary Biology
·
01 Jan 2009
·
doi:10.1186/1471-2148-9-13
2008

Nonself vegetative fusion and genetic exchange in the arbuscular mycorrhizal fungus Glomus intraradices
New Phytologist
·
24 Dec 2008
·
doi:10.1111/j.1469-8137.2008.02726.x

Multilocus genotyping of arbuscular mycorrhizal fungi and marker suitability for population genetics
New Phytologist
·
15 Oct 2008
·
doi:10.1111/j.1469-8137.2008.02602.x

Genetic diversity and host plant preferences revealed by simple sequence repeat and mitochondrial markers in a population of the arbuscular mycorrhizal fungus Glomus intraradices
New Phytologist
·
20 Feb 2008
·
doi:10.1111/j.1469-8137.2008.02381.x
2007

Gene Copy Number Polymorphisms in an Arbuscular Mycorrhizal Fungal Population
Applied and Environmental Microbiology
·
01 Jan 2007
·
doi:10.1128/AEM.01574-06
2005

Genetic variability in a population of arbuscular mycorrhizal fungi causes variation in plant growth
Ecology Letters
·
22 Nov 2005
·
doi:10.1111/j.1461-0248.2005.00853.x
